Zika reached Northeast Brazil one and a half years before it was recognized as a public threat, according to two articles written by different groups and published online in the journal Nature on May 24, 2017. The virus spread quickly, taking advantage of Aedes aegypti mosquitoes and a human population that had virtually no immunity to it. It also disguised itself as dengue and chikungunya, illnesses that present similar symptoms. Researchers working in parallel relied on different resources but shared a common goal: to monitor the evolution of the viral genome not only to understand what was happening but also to predict new outbreaks and keep diagnostic methods up to date.
A portion of these findings come from ZiBRA, the Zika in Brazil Real-Time Analysis Project. Using a mobile lab equipped with a genome sequencing device that fits in the palm of the hand, an international team has been investigating the path of the Zika virus ever since it landed in Brazil and began spreading across the Americas (see Pesquisa FAPESP Issue No. 239). They have made the entire genome sequence available to other research teams, broadening the scope of the work. “By combining epidemiological and genetic data, we were able to see that Zika was silently circulating in all regions of the Americas at least a year before its presence was first confirmed, in May 2015,” says Portuguese-born Nuno Faria, a physician-scientist at the University of Oxford and the first author of the article that describes the results of the 2016 monitoring efforts.
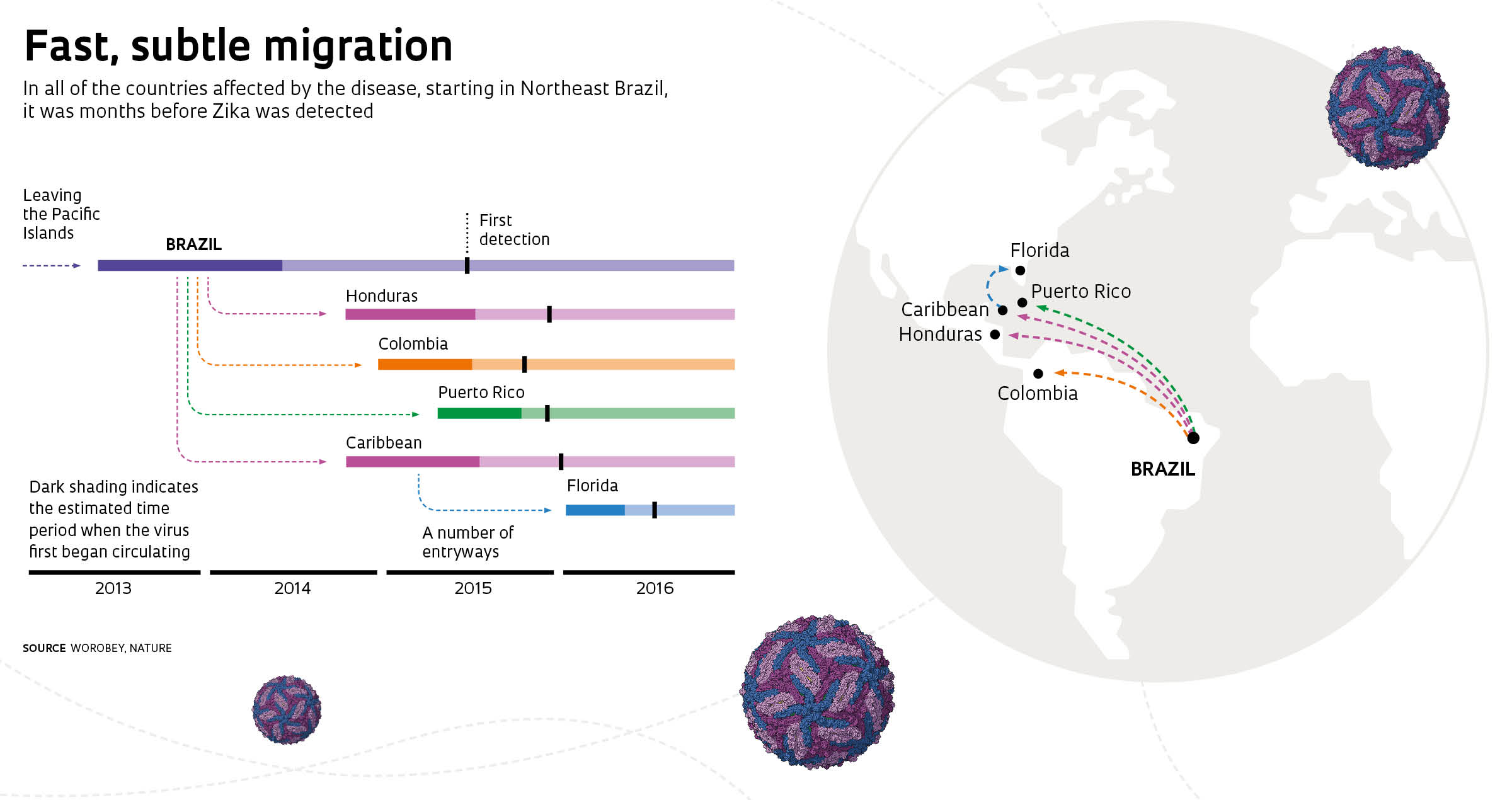
According to Faria, the virus was introduced into Northeast Brazil in February 2014. There was likely some low-level transmission in the region that year but nothing very significant. “The major outbreak most likely took place in 2015, concurrent with the dengue outbreak. Zika spread from Northeast Brazil to the Southeast region [where it first struck Rio de Janeiro] and to the Caribbean and other countries in South and Central America, eventually reaching Florida,” he says.
The ZiBRA Project based its conclusions on the analysis of 254 whole genome sequences of the pathogen, 54 of which were sequenced for the study published in Nature. Most of this new genetic data was obtained using MinION, a portable sequencer weighing less than 100 grams that is manufactured by Oxford Nanopore Technologies. The protocols that enabled the use of this technology to sequence the Zika virus were designed as part of the ZiBRA Project and eventually gave rise to a second article, published online by Nature Protocols on the same date. This method was adapted to the virus circulating in Brazil at the Institute of Tropical Medicine of the University of São Paulo (IMT-USP), under the leadership of epidemiologist Ester Sabino and in partnership with collaborators from the University of Birmingham, in the United Kingdom. “Our intern Ingra Claro tested the samples according to instructions from Joshua [Quick, first author of the article on the method] in order to obtain enough viral RNA,” Sabino explains.
The greater the number of sequences generated, Sabino adds, the easier it is to understand when the virus entered Brazil, how it spread across the continent, and mainly, how it is evolving. This analysis is possible thanks to the molecular clock technique, which evaluates the accumulation of mutations to certain genes. These changes occur at a relatively constant rate and the genes serve as timers that indicate how long it has been since the viral isolates diverged.
Set up on a bus, the mobile lab visited the states of Rio Grande do Norte, Paraíba, Pernambuco, and Alagoas in 2016
Laboratory on the road
“The idea for the project emerged in 2016 when part of the group published the first epidemiological and genetic findings on Zika in the Americas in Science. We had sequenced seven viral isolates, but the number of samples wasn’t enough to provide a broad notion of the virus’s diversity on the continent,” says geneticist Luiz Carlos Alcântara, of the Oswaldo Cruz Foundation (Fiocruz) in Bahia.
The ZiBRA Project was approved as a result of a call for proposals issued jointly by three British research funding agencies: the Medical Research Council, the Newton Fund, and Wellcome Trust. Researchers from a number of institutions have joined in the initiative, including Fiocruz, the Evandro Chagas Institute, Brazil’s Ministry of Health, USP, and the Birmingham and Oxford universities.
During 2016, a lab installed on a bus visited Central Public Health Laboratories (LACEN) in the states of Rio Grande do Norte, Paraíba, Pernambuco, and Alagoas. Other leaders of the initiative, alongside Alcântara, Faria, and Sabino, were researchers Nicholas Loman, of the University of Birmingham, Oliver Pybus, of the University of Oxford, and Marcio Nunes, of the Evandro Chagas Institute in Pará. “At each Central Lab, we analyzed 300 to 400 blood samples from patients with suspected Zika, performing a total of 1,330 tests. We did real-time diagnosis, and when the results were positive, the virus’s genetic material was sequenced,” reports Alcântara.

Ricardo Funari
Jaqueline Goes de Jesus, of Fiocruz, and Nuno Faria use a handheld MinION to monitor Zika in João Pessoa, ParaíbaRicardo FunariWith support from fixed laboratories at Fiocruz, in Salvador, Bahia, and at IMT-USP in São Paulo, the team also analyzed samples from Southeast Brazil and the state of Tocantins. Collaborators in the United States sequenced the genomes of four viral isolates from Mexico and five from Colombia. “The analyses showed that, up until 2016, the viruses found in the various regions of Brazil and neighboring Latin American countries were not yet displaying much diversity,” says Alcântara.
According to Alcântara, the African strain reached Asia shortly before 2007, when it triggered the first epidemic in Micronesia. Further outbreaks were recorded in the Philippines (2012) and French Polynesia (2013 and 2014). Zika next struck Brazil, which has reported the largest number of cases to date, surpassing 200,000 by December 2016. “The virus has changed a lot since leaving Africa. Seven to 10 years from now, there will probably be much more diversity here in the Americas. We have to maintain genomic surveillance so we are prepared if a new outbreak comes,” advises Alcântara.
In addition to assisting Central Labs to diagnose hundreds of suspected Zika cases, ZiBRA researchers also trained teams to conduct genomic surveillance using MinION. The second stage of the project is now underway, which involves monitoring the dengue, chikungunya, and yellow fever viruses, in addition to Zika. “We’re going to set up a fixed lab in Manaus to analyze Central Lab samples from Amapá, Acre, Amazonas, Roraima, and Rondônia. In October 2017, we’re taking a mobile lab to the Central-West, and in March 2018, we’ll head to the Southeast,” says Alcântara.
The risk that researchers will be caught by surprise has decreased thanks to genetic surveillance and collaborative networks
Monitoring from a lab bench
Real-time monitoring is also the focus of the efforts of another international group, although it performs its genetic analyses in fixed laboratories. The team used some of the same procedures but combined the protocols developed for MinION with the use of more powerful devices. “The focuses were complementary,” says geneticist Bronwyn MacInnis, of the Broad Institute in the United States. The study led by her in partnership with her colleague Pardis Sabeti involved sequencing 110 Zika genomes from samples collected in 10 countries. The fact that the two teams took different paths but reached similar conclusions lends support to their interpretations and validates the new sequencing techniques they have developed, which in turn provides new strategies for tracking epidemics.
One challenge faced by both groups is that the Zika virus typically presents a low viral load or viremia. “By the time the patient seeks help, the infection is already going away,” says MacInnis. In her earlier experience, during the Ebola outbreak that hit Africa in 2015, she found 1,000 to 10,000 times more viral copies in samples drawn from patients. Despite this difference, training with Ebola was the preliminary step that enabled MacInnis and her collaborators to plunge into researching the Zika epidemic without even having time to catch their breath. “That was the first time that genetic monitoring of an epidemic occurred in real time,” she says. Prior to that, samples had to be collected from patients and cultured in a laboratory in order to obtain enough virus for testing. The problem is that not everything present in the sample will grow in culture, and as a result, a great deal of diversity is lost in this process. The novel feat accomplished by the newer techniques was managing to perform genetic analyses directly on blood drawn from patients, using techniques that capture the genetic material—RNA in the case of Zika—directly from the sample.

Ricardo Funari
In Natal, Marta Giovanetti, of Fiocruz, prepares sequencingRicardo FunariWhen the epidemic broke out in Brazil and other countries in South America and the Caribbean, MacInnis looked for partners in Brazil with whom to exchange knowledge. She forged what she describes as a rich collaborative relationship with Fernando Bozza, Thiago Souza, and Patricia Bozza, all at Fiocruz in Rio. “They brought an understanding of how the disease was progressing and how it interacted with the viruses that cause dengue and chikungunya,” she says.
The group from Rio de Janeiro had already acquired an epidemiologic understanding of dengue by working with hospitals and conducting systematic surveillance of the disease. “Zika required a lot of work in the beginning because of the low viremia,” recalls Fernando Bozza. After successfully developing a quick diagnostic test, his team began collecting samples and working to improve their success in extracting RNA, which required the enforcement of more rigorous methods of collecting and storing the material.
Bozza believes that the delay in the identification of Zika after it entered Brazil underscores the importance of genetically monitoring relevant diseases that have already been detected on other continents. “By the time we identified the problem, an epidemic was already underway.” If researchers understand the evolution of the virus and possess surveillance techniques, they can develop strategies for detecting diseases faster.

Ricardo Funari
Examining mosquitoes captured in NatalRicardo FunariGenetic data indicate that it was months before the first cases of the virus were detected in Puerto Rico, Honduras, Colombia, and areas of the Caribbean and the United States. Its subtle circulation allowed Zika to move from the Caribbean into the United States, as reported in a fourth article, also published online in Nature on May 24, 2017. However, Florida is the only state where conditions are suitable for Aedes aegypti to remain year-round, allowing the virus to spread. The disease has thus been confined to that state, particularly to the Miami region, a destination for large numbers of visitors from abroad. “The virus was introduced many times. It wasn’t an isolated event,” asserts MacInnis, coauthor of the paper. “Understanding how this happened is important in coordinating our efforts to control the vector and protect entry routes.”
MacInnis knows that while there may be a break in the action now, before summer comes to Florida, other epidemics will follow. For the time being, the Zika virus still presents a bit of a mystery—it circulated less than expected during Brazil’s previous summer. “We’re going to continue monitoring to try to understand how the virus advances,” says Sabino, whose team has been tracking donated blood samples at four major blood banks in São Paulo. “We’re learning ways to put together research groups that can respond quickly in an emergency.” The risk that researchers will be caught by surprise has decreased as genetic surveillance and collaborative networks have acquired new knowledge. These simultaneous publications drive home the fact that addressing epidemics depends vitally on teamwork that draws in experts from different fields. The two groups working in parallel were aware of this and therefore did not close themselves off. “We kept in touch throughout the process, comparing our findings,” says MacInnis.
Project
Complete genomes for dengue virus: viral genetic diversity among seropositive first-time blood donors and recipients in two blood centres in Brazil (No. 12/03417-7); Grant Mechanism Scholarships in Brazil – Doctorate; Principal Investigator Ester Cerdeira Sabino (USP); Grantee Antonio Charlys da Costa; Investment R$145,246.14.
Scientific articles
FARIA, N. R. et al. Establishment and cryptic transmission of Zika virus in Brazil and the Americas. Nature. v. 546, i. 7658, p. 406-10. June 15, 2017.
GRUBAUGH, N. D. et al. Genomic epidemiology reveals multiple introductions of Zika virus into the United States. Nature. v. 546, i. 7658, p. 401-05. June 15, 2017.
METSKY, H. C. et al. Zika virus evolution and spread in the Americas. Nature. v. 546, i. 7658, p. 411-15. June 15, 2017.
QUICK, J. et al. Multiplex PCR method for MinION and Illumina sequencing of Zika and other virus genomes directly from clinical samples. Nature Protocols. v. 12, i. 6, p. 1261-76. May 24, 2017.