
steve gschmeissner / science photo libraryNeuron projections detect odors at the back of the nosesteve gschmeissner / science photo library
Lush tones of fruits and flowers, especially violets; vegetable-like and slightly spicy with a woody aroma; slightly fruity, with a hint of earthy minerals, with aromatic nuances of leather and chocolate. Some specialized descriptions of wines seem like a wild stretch of the imagination, but they are also evidence of a keen sense of smell. And the way the nose can detect these subtleties is second-to-none in this regard. The three-dimensional organization of genetic material seems to be responsible for the unique ability of each olfactory neuron to produce only one type of receptor for odorant molecules, according to a recent study by Bettina Malnic, a biochemist at the Chemistry Institute of the University of São Paulo (IQ-USP). “Genomic DNA is not scattered randomly within the cell like noodles in a bowl of soup,” says Malnic. Like other studies in recent decades, her new study makes it clear that the sense of smell is even more complex than the sommeliers’ descriptions, and to understand it, we need to think outside the box.
Olfactory neurons have a peculiarity in relation to other cells of the body, which have a wide range of receptors on the surface to recognize the molecules surrounding them. Of the approximately 1,000 genes in mice harboring the code for odorant receptors (approximately 400 in humans), only one is active in a given neuron. And what’s more: only one of the two copies of the gene, or alleles, is active. This specialization is essential for mapping smells in the brain—all neurons whose surfaces are marked for a certain type of receptor send impulses to the same region of the brain, which will recognize the pertinent aroma. Therefore, the complex bouquet of a glass of wine activates a number of different receptors which, in turn, affect different specialized areas in the brain. This olfactory mapping of receptors and how their impulses are received and organized in the brain earned the Nobel Prize in Physiology or Medicine for two Americans, Linda Buck and Richard Axel, in 2004, and has been occupying Malnic throughout her career.
A few years ago, she discovered that the odorant molecules are contained in more than one receptor, even though these may be very specific (see Pesquisa FAPESP Issue No. 155). They are molecules with multiple tips, each with a different fit. Because it is connected to more than one neuron, each odorant molecule is able to activate more than one area in the brain. It is this complex code that allows a large aromatic repertoire to be detected, by combining the action of multiple receptors.
For humans, who are not known for their keen sense of smell, it was believed that this repertoire consisted of about 10,000 odors. Recently, however, a study by researchers at Rockefeller University in the United States has greatly increased this estimate. The U.S. researchers made a series of mixtures with 10, 20 or 30 components from a repertoire of 128 odorant molecules, and tested the ability of well-trained nose volunteers to distinguish among them, according to an article published in the March 2014 issue of Science. Based on these results, a series of calculations led to the figure of 1 trillion olfactory stimuli. Malnic does not believe that this number should be taken quite so literally, but it is important for being several orders of magnitude higher than the previous estimate. “It goes against the notion that smell is not important for humans,” she says.
What was not known until now was the silencing mechanism of the inactive genes in each of the neuron projections on the surface of the epithelium at the back of the nose. “We are trying to understand how the olfactory neuron performs the feat of expressing only one of the two copies of a single gene, in such an efficient manner.” To understand the regulation of the genes responsible for constructing the receptors for odor molecules, Malnic is analyzing the nucleus of the neurons as a structure in which the genetic material has a precise spatial organization. “The nucleus is not like a bowl of soup with its ingredients scattered about, but rather made up of compartments, each with a different role to play.” she says.

bettina malnic / uspIn the nasal epithelium of transgenic mice, neurons in which the P2 gene is active appear in fluorescent greenbettina malnic / usp
Architecture
In mouse cells, the USP group used a technique known as three-dimensional immuno-DNA Fish, which allowed them to locate the olfactory receptor genes within the nucleus. “It’s as if we divided the nucleus into slices, which we can then join and obtain a three-dimensional image,” explains Malnic. The work, published in the February 2014 issue of PNAS, was largely done by Lucia Armelin-Correa, a biologist, during her post-doctorate in Malnic’s laboratory. She used a high-resolution microscope to view the structures of the nucleus of the neurons. What she saw was an unexpected organization of regions where DNA is wound in a more compact way—the heterochromatin—where gene functioning is inhibited, and active areas—the euchromatin—where genetic material has more physical space for its biochemical reactions. Armelin-Correa, Malnic and other members of the laboratory detected a peculiarity in the olfactory neurons: the heterochromatin is condensed into a sphere near the center of the nucleus, rather than at several smaller and peripheral points, as in other cell types.
The result is consistent with that of the group led by Stavros Lomvardas, of the University of California, San Francisco (UCSF), in an article published at the end of 2012 in the journal Cell. According to the U.S. group, which Malnic considers a good competitor, this three-dimensional landscape where genes and regulatory sequences are hidden, or exposed, depending on the type of cell, may be essential to determine the specific characteristics of each tissue.
With a fluorescent probe that recognizes all of the genes that encode olfactory receptors, the Lomvardas team showed that these molecular clusters as a whole are aggregated in the compartment that represses the activity, the heterochromatin. Malnic’s group did a more detailed analysis and investigated the nuclear structure with which four regions of DNA with genes for olfactory receptors, located on three different chromosomes, are associated. “For all four genes, we found that in most nuclei there was one allele next to the heterochromatin and another that was not,” says Malnic. As a control, the researchers also monitored the gene of an olfactory protein that is always active: this was associated with heterochromatin in only 20% of the nuclei examined. USP’s Malnic is cautiously optimistic that the results may explain the systematic inactivation of one allele of the genes for odor receptors.
An intriguing result was that about 45% of the fluorescent tags for one of the genes were associated with heterochromatin, and only 17% with euchromatin, where the active alleles should be. It was an indication that some of them must be linked to some other structure.
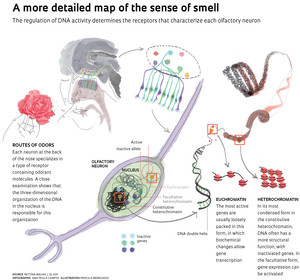 Upon further investigation, Malnic’s team saw it was necessary to analyze two types of heterochromatin to locate the two copies of each gene. The constitutive heterochromatin, concentrated in the center of the nucleus, harbors at least one of the alleles in most cells. The other is usually located next to the facultative heterochromatin, which in olfactory neurons is also concentrated in a central area of the nucleus, forming something like a hat around the constitutive. According to the gene studied, in 60% to 73% of the nuclei examined at least one allele was associated with facultative heterochromatin. As the name indicates, this structure can de-condense and change its properties, so that the alleles located there would have the potential to become active. “The mechanism of this repression is more plastic yet little studied,” says Malnic, who believes that the two types of heterochromatin work together to regulate the gene expression of odor receptors. “The distribution pattern of the two heterochromatin types indicates that something important is happening here.”
Upon further investigation, Malnic’s team saw it was necessary to analyze two types of heterochromatin to locate the two copies of each gene. The constitutive heterochromatin, concentrated in the center of the nucleus, harbors at least one of the alleles in most cells. The other is usually located next to the facultative heterochromatin, which in olfactory neurons is also concentrated in a central area of the nucleus, forming something like a hat around the constitutive. According to the gene studied, in 60% to 73% of the nuclei examined at least one allele was associated with facultative heterochromatin. As the name indicates, this structure can de-condense and change its properties, so that the alleles located there would have the potential to become active. “The mechanism of this repression is more plastic yet little studied,” says Malnic, who believes that the two types of heterochromatin work together to regulate the gene expression of odor receptors. “The distribution pattern of the two heterochromatin types indicates that something important is happening here.”
So far, the work has answered some questions and led to many others, with the possibility of broadening the perspective to the entire genome. For now, the study showed that the organization of heterochromatin and euchromatin may be different for each cell type, with a major impact on gene activity. “The neighborhoods between genes may vary according to the tissue,” says Malnic. With the help of these nuclear structures, DNA can wind itself so that genes that are far apart, when the strand is stretched out, could eventually come together and work in tandem and influence each other through the molecules they produce.
“The sense of smell is a model,” says Malnic. For her, olfactory neurons are suitable for this type of study due to their gene inactivation system. What they reveal can, she hopes, aid in understanding the regulation of genetic material in other cell types.
Projects
1. The molecular mechanisms of smell (nº 2011/51604-8); Grant mechanism Thematic Project; Principal investigator Bettina Malnic (IQ-USP); Investment R$809,219.21 (FAPESP).
2. Regulation of gene expression of olfactory receptors: study of nuclear architecture of olfactory neurons and the relative positioning of active and inactive alleles (nº 2007/57734-5); Grant mechanism Post-doctoral research grant in Brazil – Regular; Principal investigator Bettina Malnic (IQ-USP); Grant recipient Lucia Maria Armelin-Correa; Investment R$ 222,662.28 (FAPESP).
Scientific article
ARMELIN-CORREA, L.M. et al. Nuclear compartmentalization of odorant receptor genes. PNAS. V. 111, No. 7, p. 2782-87. February 18, 2014.