 Léo RamosHaving determined that the protein alpha tubulin is altered by an enzyme called protein kinase C during cell division, biomedical scientist Deborah Schechtman at the University of São Paulo Chemistry Institute (IQ-USP) wanted to know more about how this happens. She needed to find exactly where the two molecules fit together, like pieces in a jigsaw puzzle. Although the proteins in cells fold themselves into complex tangles, scientists usually turn to their linear sequences when looking for kinase binding sites. The linear strand of a protein is like a string of colored beads, each color representing a different amino acid. After failing to find what she was after, Schechtman had an inspired idea: she called biologist Paulo Oliveira at the Brazilian Biosciences National Laboratory (LNBio) and asked him to take a look at the three-dimensional structure of the protein. It didn’t take long for the molecular modeling expert to call back with the news: he’d found it.
Léo RamosHaving determined that the protein alpha tubulin is altered by an enzyme called protein kinase C during cell division, biomedical scientist Deborah Schechtman at the University of São Paulo Chemistry Institute (IQ-USP) wanted to know more about how this happens. She needed to find exactly where the two molecules fit together, like pieces in a jigsaw puzzle. Although the proteins in cells fold themselves into complex tangles, scientists usually turn to their linear sequences when looking for kinase binding sites. The linear strand of a protein is like a string of colored beads, each color representing a different amino acid. After failing to find what she was after, Schechtman had an inspired idea: she called biologist Paulo Oliveira at the Brazilian Biosciences National Laboratory (LNBio) and asked him to take a look at the three-dimensional structure of the protein. It didn’t take long for the molecular modeling expert to call back with the news: he’d found it.
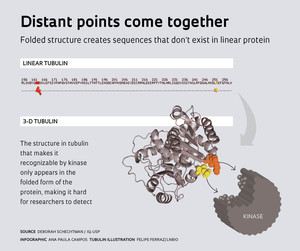
“When a protein folds itself, two far-apart areas of its linear structure can end up close together,” explains Schechtman, who says the finding – published in Science Signaling in November 2014 – is a break of paradigm. The editor of the journal says the process is similar to origami, in which paper is folded to create a recognizable structure. This is an important breakthrough for researchers in the field, who use bioinformatics software to access protein databases and compare their linear structures – their strings of colored beads – to find consensus sequences that are essential for recognition between proteins or between proteins and other types of molecules. This traditional method has been effectively used to pinpoint a great many interaction sites, but the groups headed by Schechtman and Oliveira analyzed structural models of approximately 1,000 proteins and showed that structural motifs (three-dimensional arrangements) are also a recurring characteristic. “Now, we need to develop bioinformatics tools to predict three-dimensional binding sites,” Schechtman concludes.
It’s hardly news that the 3-D structure of proteins is essential to their function, but this premise is not usually taken into account when researchers study the interactions between kinases and their substrates, by which intracellular events are triggered by stimuli originating outside the cell. In the case studied by Schechtman at USP, a stimulus activates a receptor located on the cell membrane, setting off a series of reactions that ultimately activate the enzyme protein kinase C. The enzyme then moves to the cell region where it is supposed to work. There, protein kinase C finds alpha tubulin and transfers a phosphate group to it (in a process called phosphorylation). This last interaction is where three-dimensional recognition comes in. “We showed that kinases can read Braille,” jokes Schechtman, in a reference to recognizing something by touch as opposed to reading the linear sequence of letters representing specific amino acids.
 As for the impact of the discovery, she expects to see it in the coming years as related scientific publications come out, but the quick response that only social networks can provide was enough to give her a preview. The day after publication of the paper in Science Signaling, Portuguese researcher Pedro Beltrão from the European Bioinformatics Institute (UK) posted the news on Twitter – the social network most commonly used for such announcements (see Pesquisa FAPESP Issue No. 221). “The specificity of domain-peptide interactions was already a hard problem. Now we have to think about 3D ‘linear’ motifs,” he wrote, triggering interested responses from colleagues in several countries and initiating a brief online discussion. The Twitter posts revealed that the challenge of thinking in three dimensions is not the only change, but also the larger search areas that researchers must scrutinize in their search for molecular interaction sites. “Phosphorylation was believed to be more frequent in unstructured regions of proteins,” Schechtman explains. This is not what she and her colleagues saw.
As for the impact of the discovery, she expects to see it in the coming years as related scientific publications come out, but the quick response that only social networks can provide was enough to give her a preview. The day after publication of the paper in Science Signaling, Portuguese researcher Pedro Beltrão from the European Bioinformatics Institute (UK) posted the news on Twitter – the social network most commonly used for such announcements (see Pesquisa FAPESP Issue No. 221). “The specificity of domain-peptide interactions was already a hard problem. Now we have to think about 3D ‘linear’ motifs,” he wrote, triggering interested responses from colleagues in several countries and initiating a brief online discussion. The Twitter posts revealed that the challenge of thinking in three dimensions is not the only change, but also the larger search areas that researchers must scrutinize in their search for molecular interaction sites. “Phosphorylation was believed to be more frequent in unstructured regions of proteins,” Schechtman explains. This is not what she and her colleagues saw.
Adequate interaction between kinases and proteins is crucial to human health. Imbalances in these interactions could be behind the development of cancer, inflammatory processes, and cardiovascular problems, among other illnesses. “Kinase modulators account for 25% of all efforts in the pharmaceutical industry,” says Schechtman. She believes that understanding how these enzymes interact with proteins can contribute to the design of more specifically targeted modulators than those available today.
“If we perform good basic research, it will eventually prove useful,” says the researcher from USP, defending broad funding for investigations that do not directly address a specific practical concern. She also highlights the importance of multidisciplinarity in research groups and researchers themselves. “The only reason I looked at three-dimensional structures was that I used to work in biochemistry before shifting to cell biology.” Her partnership with Paulo Oliveira began when both were sharing an office at USP’s Heart Institute (InCor). The result was a project involving students advised by both researchers – in this case, mainly PhD students Mariana Duarte and Darlene Pena and master’s degree student Felipe Ferraz –, pooling their knowledge of biochemistry, cell biology, and protein modeling.
The importance of maintaining a precise 3D protein structure was also recently demonstrated by a group led by chemist Peter Wolynes and Brazilian physicist José Onuchic at Rice University, USA. In a paper published in PNAS in August 2014, they reported evidence that natural selection applies strong pressure for maintaining the structural integrity of folded molecules, in eight families of proteins. They observed that when a mutation changes an amino acid in a part of the protein that interacts with another, the second part also changes in order to preserve the original structure. This happens even when the two segments are far apart in the protein’s linear structure. From an evolutionary perspective, this finding corroborates the role of three-dimensional structure in protein functionality.
Projects
PKC and signal transduction pathways of self-renewal and differentiation in murine embryonic stem cells (No 2010/18640-8); Grant mechanism: Regular Line of Research Project Award; Principal investigator: Deborah Schechtman (USP); Investment: R$412,740.16 (FAPESP).
Rational design of protein kinase C specific inhibitor peptides: a computational approach and experimental validation (No 2008/52695-4); Grant mechanism: Regular Line of Research Project Award; Principal investigator: Paulo Sergio Lopes de Oliveira (LNBio); Investment: R$90,016.12 (FAPESP).
Scientific articles
DUARTE, M. L. et al. Protein folding creates structure-based, noncontiguous consensus phosphorylation motifs recognized by kinases. Science Signaling. V. 7, No. 350, ra105. Nov. 4, 2014.
MORCOS, F. et al. Coevolutionary information, protein folding landscapes, and the thermodynamics of natural selection. PNAS. V. 111, No. 34, p. 12408-13. Aug. 26, 2014.

