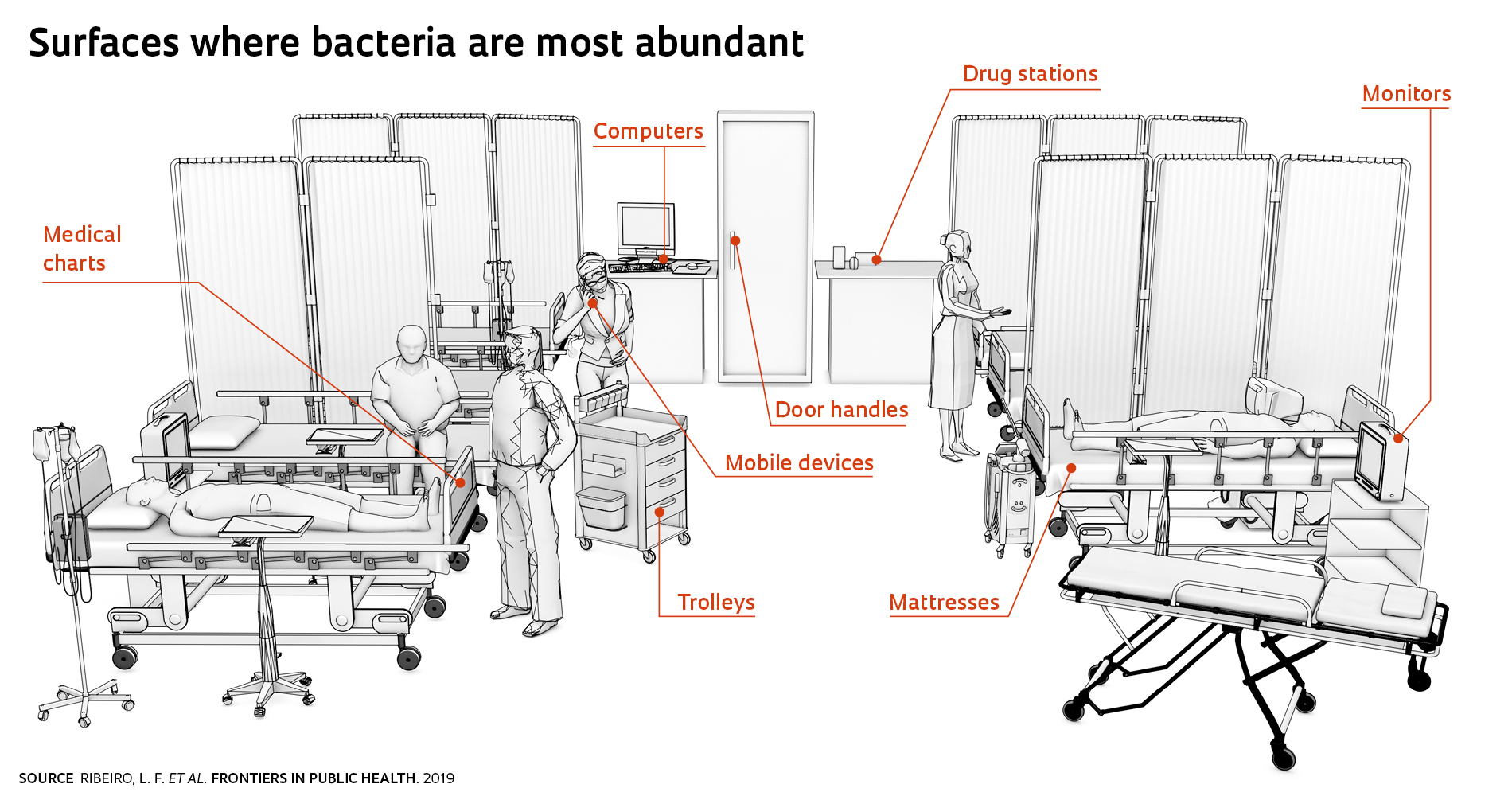
Bacteria appear to have become adapted to the environment in intensive care units (ICU), where patients with serious conditions are meant to be kept safe from infectious agents. These are the findings from a study by researchers at Hospital das Clínicas (HC) of the Ribeirão Preto School of Medicine at the University of São Paulo (FMRP-USP), a referral center in southeastern Brazil. The study identified 138 bacterial genera in the adult ICU and 160 in the neonatal ICU at HC. The microbes were found on mattresses, bed rails, furniture, computer touch screens, infusion pumps, ventilators, and mobile devices used by medical staff. The researchers found that, even after standard cleaning procedures were performed, most bacteria survived. Microbes find their way into an ICU primarily via the seemingly clean hands of medical staff, patients, and visitors. With 817 hospital beds and 105 ICU beds, HC has almost 6,000 staff and handles around 35,000 hospitalizations per year—or almost 100 per day.
Most identified bacteria are generally harmless in healthy individuals, but can cause serious infections in ICU patients whose body defenses are impaired. The risk for microbial infections is as much as 10 times higher in ICUs than in other departments of a hospital, according to the Brazilian Intensive Care Association. The most common infections are pneumonia and infections of the bloodstream and urinary tract. This is a global issue that is being tackled through continual improvement of cleaning protocols.
There are currently no benchmarks against which to determine whether the bacterial diversity found in the ICUs at HC is above or below acceptable levels. “The contamination risk for patients needs to be evaluated,” says infectious disease specialist Gilberto Gaspar, who heads the Hospital Infection Control Committee and participated in the study, published in August in Frontiers in Public Health. He argues that the amount of bacteria needed to cause an infection is unknown. “We need to revisit the methods, frequency, and effectiveness of cleaning procedures,” he says. The hospital does not disclose its ICU infection rates to the general public.
The hands of 60% of medical staff at a hospital in Uberlândia were contaminated with bacteria
At HC, two genera of bacteria, Pseudomonas and Staphylococcus, were abundant: they were found on mattresses, bed rails, inhalation pumps, door handles, cabinet doors, dressing trolleys, drug stations, and computers. “Pseudomonas are bacteria found in the soil, and are likely carried into ICUs on the shoes worn by medical staff and visitors,” says microbiologist María Eugenia Guazzaroni, a professor at the University of São Paulo’s Ribeirão Preto School of Philosophy, Sciences, and Languages and Literature (FFCLRP-USP), who led the study. They can cause respiratory and urinary tract infections, otitis, and conjunctivitis. Staphylococcus bacteria are found on human body surfaces, such as the nasal cavities, and can cause a variety of infections in ICU patients, including pneumonia and sepsis. “But people’s hands are still the primary sources of bacteria,” says biochemist Lucas Ribeiro of FFCLRP-USP. Microbe transmission could be mitigated if staff and visitors adhered more conscientiously to hygiene standards.
The level of bacterial resistance to routine disinfection procedures was startling—cleaning with wipes soaked in broad-spectrum antimicrobial solutions was insufficient to completely eliminate microorganisms in the ICUs. At both units, cleaning reduced the number of bacterial genera by only 20%. But cleaning effectiveness was not consistent across different surfaces, suggesting that the variation in bacterial diversity before and after cleaning could be partly explained by a lack of standardized procedures. Biocide rotation, a practice adopted in hospitals in Europe and the US, could reduce the ability that microbes have to adapt to cleaning protocols.
A broad-spectrum method
The diversity of bacteria in ICUs at the Ribeirão Preto hospital was determined using a technique called metagenomics. This approach consists of large-scale analysis of genetic material using 16S rRNA sequencing to identify bacteria. The method is capable of identifying unculturable bacteria down at least to the genus level.
Only 2.5% of all bacterial species are culturable, according to researchers at the Graz University of Technology and the Medical University of Graz, Austria. Using this sequencing method, which can theoretically detect all microorganisms, they found 76 bacterial genera with at least 1% abundance in 36 surface samples taken in ICUs at Graz Medical University. Sample material was collected from medical devices such as respirators, ultrasound devices, and endoscope tips, and from workspaces including doctors’ rooms and procedure rooms, the visitor’s entrance, and patient rooms. According to a March 2013 article in Scientific Reports, the most prevalent bacteria—and the primary sources of infections—were those of the genera Pseudomonas, Propionibacterium, and Burkholderia.
Similarly, the genera Bacillus, Staphylococcus, and Pseudomonas were the most abundant (47% of the samples) in the adult ICU at HC, whereas Bacillus, Propionibacterium, and Staphylococcus were the most prevalent (40%) in the neonatal ICU. Microorganisms of these genera, which are also found in hospital ICUs in the US and Europe, can survive for months on dry surfaces.
On people’s hands and mobile devices
In another study, biologist Marina Aparecida Soares, of the Federal University of Uberlândia, Minas Gerais, southeastern Brazil, collected 26 surface samples from the hands of nursing staff and physicians in the adult ICU and 25 from the neonatal ICU at the Dr. Odelmo Leão Carneiro Municipal Hospital and Maternity Center, in Uberlândia. The hands of 60% of participants were contaminated with bacteria, including Pseudomonas aeruginosa, as reported in an article in the July-September issue of Revista de Epidemiologia e Controle de Infecção.
The mobile devices of staff in the adult ICU at HC were teaming with bacteria of the genera Acinetobacter, Sphingomonas, and Brevundimonas, which are common in humid environments, while the bacteria most abundant on devices in the neonatal ICU were of the genera Fusobacterium, Neisseria, Rothia, Granulicatella, and Streptococcus, which are among the microbiota found in the nose and on the skin. “A recommendation which is rarely followed is never to allow mobile devices into ICUs,” says Mayra Menegueti, a nurse who serves on the infection control committee at HC.
Infectious disease specialist Denise Brandão de Assis, technical director of the Hospital Infection Division at the São Paulo State Department of Health’s Epidemiological Surveillance Center (CVE), says ICU hygiene is a persistent problem—and not just in Brazil. “People believe the ICU environment is clean, when in fact it isn’t,” she says. The problem is a serious one, but can be mitigated. A CVE survey of 652 private, charity-run, and public hospitals in São Paulo State reported a near-steady drop in urinary tract infection (77% reduction), pneumonia (68%), and bloodstream infection (47%) rates in ICUs from 2004 to 2018, a result of improved hygiene protocols.
Project
Novel approaches to improve functional screening of biocatalysts in metagenomic libraries (nº 15/04309-1); Grant Mechanism Junior Researcher; Principal Investigator María Eugenia Guazzaroni (USP); Investment R$1,360,080.65.
Scientific articles
RIBEIRO, L. F. et al. Microbial community profiling in intensive care units expose limitations in current sanitary standards. Frontiers in Public Health. Vol. 7, a240, pp. 1–12. Aug. 28, 2019.
OBERAUNER, L. et al. The ignored diversity: complex bacterial communities in intensive care units revealed by 16S pyrosequencing. Scientific Reports. Vol. 3, no. 1413, pp. 1–12. Mar. 11, 2013.
SOARES, M. A. et al. Microrganismos multirresistentes nas mãos de profissionais de saúde em unidades de terapia intensiva. Revista de Epidemiologia e Controle de Infecção. Vol. 9, no. 3, pp. 1–12. July–Sept. 2019