Beginning this month, eight Brazilian public health centers specializing in rare diseases will make the first drug capable of alleviating the symptoms of spinal muscular atrophy (SMA) available to all children with the genetic condition. This type of atrophy leads to progressive loss of muscle strength and, in severe cases, early death. Approved for clinical use in the United States in 2016, and in Brazil in 2017, the drug nusinersen—marketed by American lab Biogen under the name Spinraza—has improved motor ability in 40% of children treated, according to data published in 2017 in the New England Journal of Medicine. The drug modifies the functioning of one gene and increases the production of SMN protein, which is essential for the survival of spinal cord cells that transmit brain commands to muscles.
Injected under the membranes that protect the spinal cord, nusinersen is one of the most expensive medicines in the world. When released, the six doses given during the first year of treatment cost US$750,000 in the United States. From the second year onwards, the number of applications and the cost of the lifelong therapy fall by half. In Brazil, the Unified Health System (SUS) has offered the drug since April for cases that manifest during the first six months of life and, from now forward, for cases that begin after six months. There are 300 to 400 children born with SMA each year throughout the country.
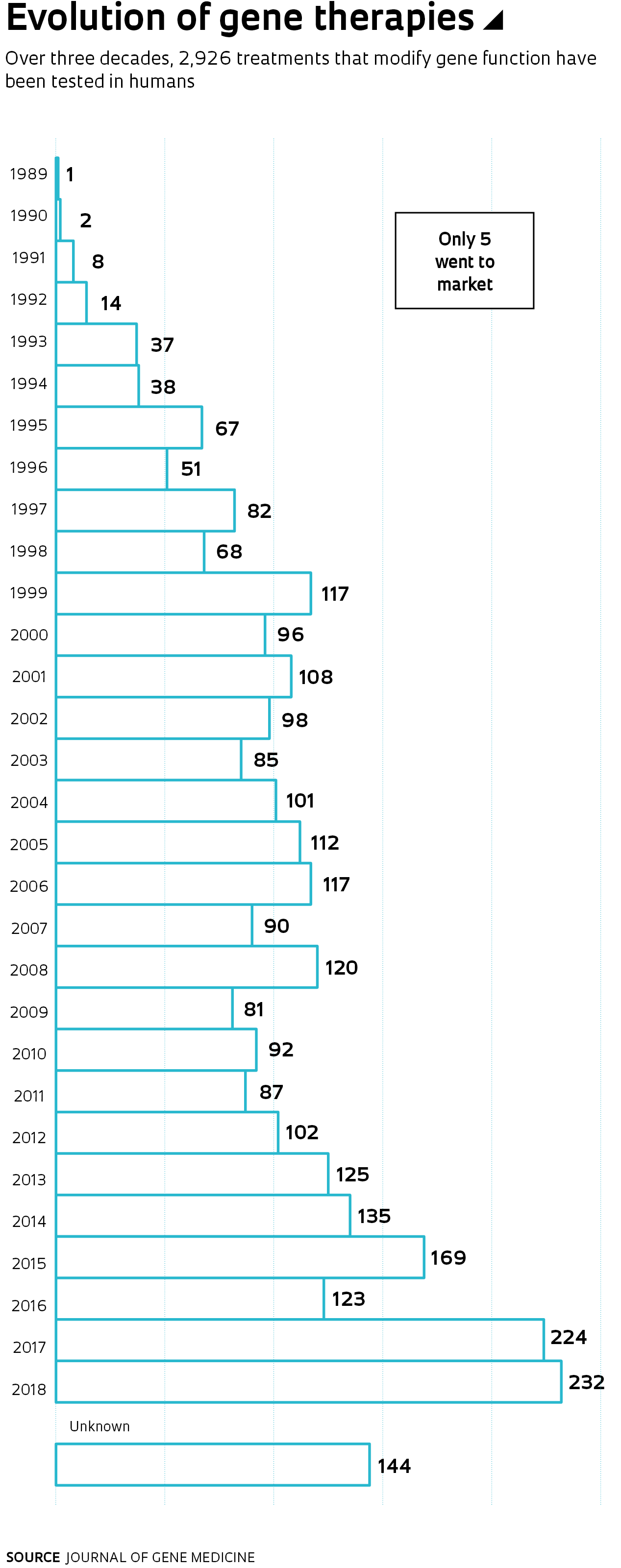
Nusinersen is a member of a new class of compounds. These drugs arose as a consequence of sequencing the human genome, which transformed molecular biology and was a frequent subject in the pages of Pesquisa FAPESP during the 20 years it’s been in print. The magazine has published at least ten cover articles regarding various genome projects and their results, as well as dozens of smaller reports. Defining the order of the 3.3 billion nitrogenous bases (adenine, A; thymine, T; cytosine, C; and guanine, G) of the human genome paved the way for faster and more accurate analysis of its genes, which in turn improved and lowered the cost of diagnosing genetic diseases. This also led to innovative treatments, some with the potential to cure. These new therapies, however, continue to be inaccessible due to exorbitant costs.
“The sequencing of the human genome has allowed important advances in the diagnoses of rare diseases,” says geneticist Lygia da Veiga Pereira, from the University of São Paulo (USP). These are diseases that are caused by changes in a single gene (monogenic), and they are generally severe. Each disease by itself only strikes a fraction of the world’s population, ranging from one in every thousand to one in every 100,000 people. Together, they affect almost 6% of the world’s population, a proportion similar to that afflicted with diabetes (8.5%). By 2000, when an international public sequencing consortium competed with the company led by US geneticist John Craig Venter to complete the task of reading and ordering the chemical letters of the human genome, 1,900 monogenic diseases were known. Today, alterations in 4,147 genes associated with 6,499 diseases have been mapped, according to the database of the Online Mendelian Inheritance in Man (OMIM).
Advances in sequencing techniques and the evolution of bioinformatics have made it possible to compare the genomes of healthy individuals with those with various diseases, and to identify the cause of monogenic conditions, which has not yet been viable for more complex diseases involving multiple genes (polygenic). “This knowledge was essential for improving identification and treatment, as well as prevention through family genetic counseling,” explains geneticist Mayana Zatz, coordinator of the Human Genome and Stem Cell Research Center (HUG-CELL) at USP, one of the Research, Innovation, and Dissemination Centers (RIDC) funded by FAPESP. At HUG-CELL, a single test detects altered genes associated with nearly 6,700 diseases (neuromuscular disease, hereditary cancers, autism, and others).
Identifying the cause of genetic disorders improves quality of life by allowing physicians to select the most effective remedies to alleviate symptoms and avoid drugs that aggravate them. It also helps to prepare family members and caregivers for how the condition will develop. There is one other priceless benefit, adds medical geneticist Iscia Lopes Cendes, coordinator of the Molecular Genetics Laboratory of the University of Campinas (UNICAMP) and a researcher at Brainn, another FAPESP-funded RIDC. “Genetic testing often gives a definitive diagnosis for these serious illnesses and reduces parental distress,” she explains.
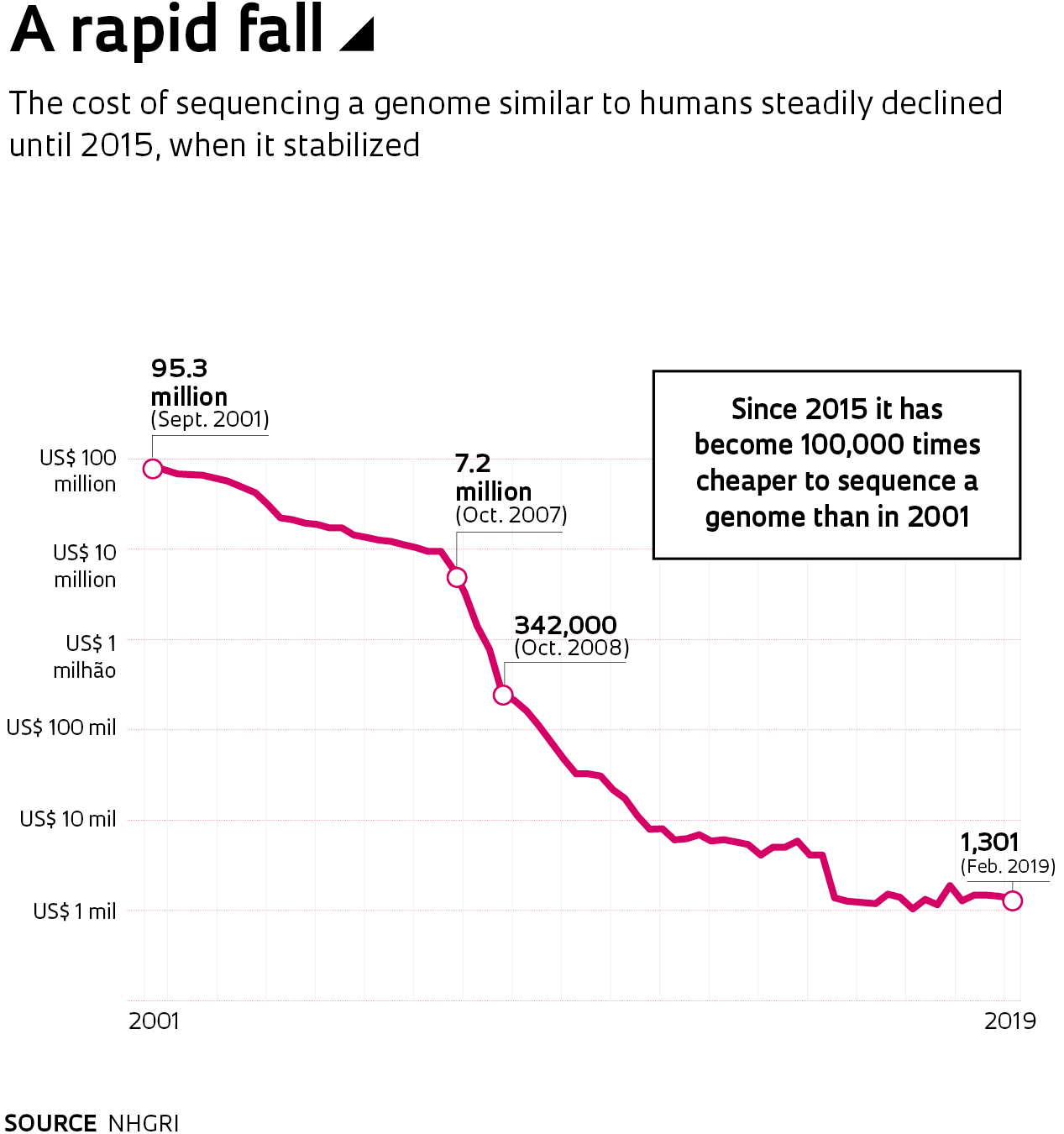
When the first version of the human genome was published in 2001, there was excessive optimism among many researchers, a feeling that reverberated through the media, raising expectations that were difficult to live up to. At the time, American geneticist Francis Collins compared the genome to a book that chronicled the journey of our species through time. Then director of the National Human Genome Research Institute (NHGRI) in the United States that coordinated the public sequencing consortium, Collins added: “It’s a transformative textbook of medicine, with insights that will give healthcare providers immense new powers to treat, prevent, and cure disease.”
This hyperbolic tone contrasted with the restraint used in scientific articles reporting the accomplishment, one published on February 15, 2001, in the journal Nature by the Collins consortium and another in Science on the next day by the Venter team. In speaking to peers, Collins’s group was cautious. They stated there would be long-term consequences for medicine and ended the article by saying, “We must set realistic expectations that the most important benefits will not be realized overnight.”
In Science, Venter and his collaborators wrote: “The sequence is only the first level of understanding the genome. All genes and their control elements must be identified; their functions, in concert as well as in isolation, defined; their sequence variation worldwide described; and the relation between genome variation and specific phenotypic characteristics determined.”
Science, as they knew, is not fast. “Over these nearly 20 years, a lot has progressed, but we haven’t yet achieved the applications that many imagined,” says Cendes.
At the doctor’s office
Advances in sequencing technologies and bioinformatics data-analysis strategies were essential so that medicine could, almost two decades later, begin to use genomics knowledge in clinical practice. “Only recently have some areas of medicine moved from a contemplative stance to a more active position,” says pediatric neurologist Fernando Kok, a researcher at the USP School of Medicine (FM-USP) and medical director at Mendelics, a company specializing in personalized genetic diagnostics. He believes there will soon emerge a wave of gene therapies but that access to them will be limited due to cost. “Increasing access will be a problem for healthcare managers,” he warns.
One engine of progress in genomics has been the enhancement of sequencing technology. In the mid-1970s, when Allan Maxam and Walter Gilbert in the United States, and Frederick Sanger and Alan Coulson in England developed the first two strategies for sequencing DNA, the process was slow and laborious. Gilbert and Sanger shared the 1980 Nobel Prize for Chemistry with biochemist Paul Berg. It took an entire day to identify the order of a few hundred base pairs of DNA. Only a decade later, automated devices that employed the Sanger method emerged and were used in the Human Genome Project.
More precise, this technique sequences only one short strand of DNA at a time, up to 900 bases. Copies are produced with an increasing number (1, 2, 3…) of bases. Only one base (A, C, T, or G) is added to each copy, and the last base is always marked with a fluorescent dye (green for A; blue for C; red for T; and yellow for G). When copies are finished, they are divided according to size. Since the last base of each copy is known, the original sequence can be reestablished. The Sanger method is still used today to sequence isolated DNA molecules, although in most applications it has been replaced by a faster and cheaper technique, known as next-generation sequencing (NGS), which can identify the order of millions of DNA bases simultaneously. In addition to the two methods above, which have been adopted in clinical laboratories, there is a third technique used in research. Single-molecule, real-time sequencing (SMRT), uses a laser source to illuminate each base—labeled with a fluorescent dye—as it is added to a DNA strand being copied.
The cost of the task dropped from US$100 million in 2001 to about $1,000 in 2015, according to NHGRI calculations. That figure remains stable, although companies are working to reduce the price of genome sequencing—or at least the price of sequencing the exome, the part that contains the 24,000 protein-coding genes—to only hundreds of dollars.
“It needed to get to the point that the techniques became very cheap, and we became good enough at interpreting the data, for this technology to become available in medical practice,” says Cendes. One study she headed with medical geneticist Antonia Marques de Faria, also from UNICAMP, helped promote the approval of a new genetic test, exome sequencing, which is used for diagnosing intellectual disability and was incorporated into the SUS system in March.
With its variety of clinical manifestations, intellectual disability is regarded as a group of rare diseases with difficult clinical diagnosis. Their various forms, together, afflict 1% to 2% of the population and affect, to varying degrees, learning, social interaction, and the capacity for self-care. The current diagnosis within the SUS system is done by karyotype testing (analysis of chromosomes, the structures in which genes are packaged), and by DNA microarray, a technique that analyzes repetitions in the genome and is still rarely available. The first method identifies the cause in 3% of cases, the second in up to 20% of cases. However, exome analysis works almost 40% of the time. Regarding the cost-benefit ratio, opting for exome analysis appears to be beneficial, according to a study by Joana Prota, a doctoral student supervised by the UNICAMP researchers.
Common diseases
While genomics has advanced the diagnoses of causes of rare diseases, it is still somewhat lacking with regard to the most common diseases such as diabetes, cardiovascular problems, psychiatric diseases, and many forms of cancer, which are important from a public health standpoint due to the large number of people affected. These are complex and multifactorial diseases that result from the activities of dozens to hundreds of genes that interact with each other and with the environment. For this reason, to date, no single gene has been found that plays a major role in the onset of hypertension, a problem that affects about one-third of the world’s adult population. Illness caused by alteration in a single gene is rare. The same is true of diabetes, psychiatric disorders, and various cancers.
In complex diseases, the contribution of each gene is small. The effect of one gene can only be quantified by comparing a large number of genomes, as is beginning to be done in England, the United States, and China, where there are projects to sequence the genetic material of up to one million people. Still, what is found in these countries may be valid only for European or Asian populations. In an article published this March in the journal Cell, University of Pennsylvania geneticist Giorgio Sirugo and two US collaborators state that genome-wide association studies designed to identify variants associated with complex traits, or with the risk of developing diseases, are focused on only a few populations: 52% were done with Europeans and 21% with Asians. According to the researchers, studying groups from other origins is important because “patterns of genetic variation between populations can affect the risk of developing disease and the effectiveness and safety of treatments.”
In Brazil, studies evaluating population genomics are still rare. At HUG-CELL, Zatz’s team conducted the exome analysis of approximately 1,500 São Paulo residents over 60, looking for protective gene variants. The Brazilian Initiative on Precision Medicine (BIPMED), coordinated by CENDES, pioneered the open data sharing of genomes of almost 900 individuals (350 of whom were healthy, representing the general population). At the A.C. Camargo Cancer Center in São Paulo, researchers recently sequenced the genome of 300 patients with stomach cancer. And at USP, Lygia Pereira currently plans to obtain genomic data from hundreds of thousands of Brazilians in order to characterize the genetic variations of the populace.
So far, however, genomic analyses can at best only show associations between the occurrence of certain genetic variants and the risk (predisposition) of developing a particular health problem. “For diabetes and obesity, for example, the contribution of these studies is still small, with the potential for more effective treatments in the medium term,” comments USP endocrinologist Alexander Jorge, a specialist in genetic diseases.
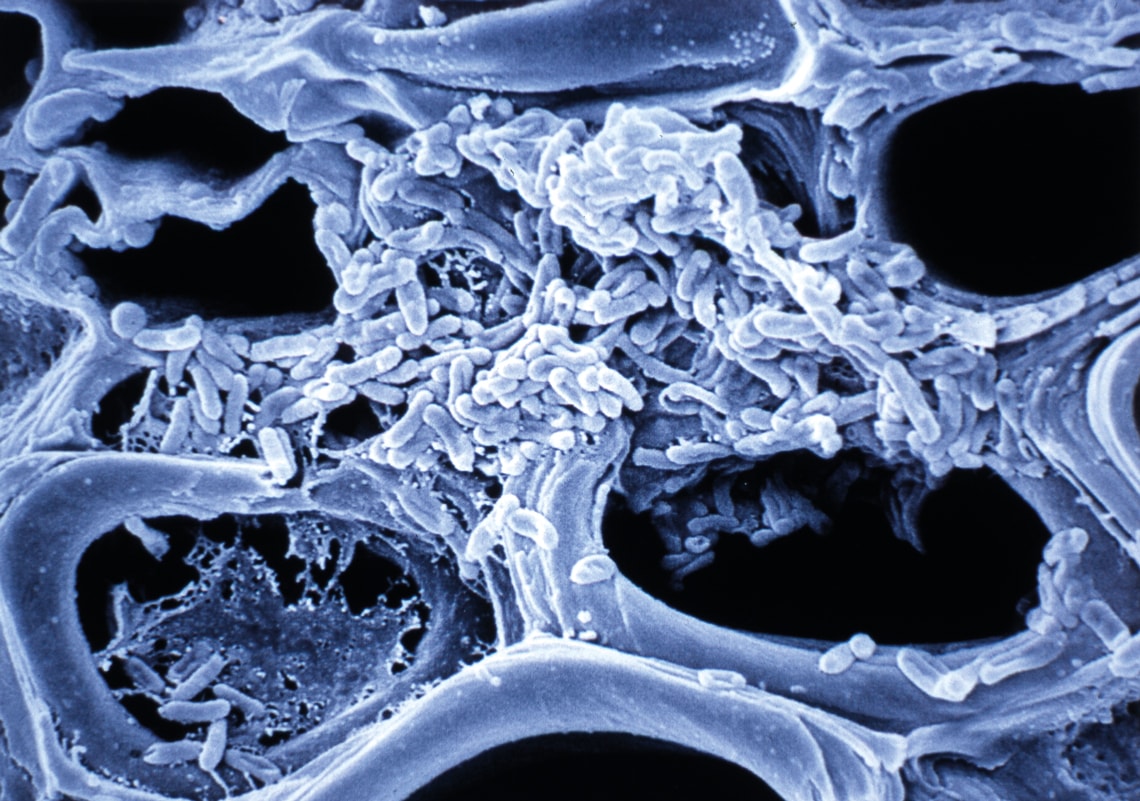
Stem veins of an orange tree, blocked by a colony of Xyllela fastidiosa bacterium, the first phytopathogen to have its genome sequenced
Despite these limitations, information on altered genes obtained from the human genome, and the projects that followed it, has aided the diagnosis and treatment of many of the nearly 200 known cancers. “In oncology, the genetic characteristics of tumors have been used to identify the type of cancer and its aggressiveness. They also allow us to monitor the evolution of the disease and the response to treatment,” says geneticist Anamaria Camargo, coordinator of the Molecular Oncology Center at the Syrian-Lebanese Institute for Education and Research (IEP), in São Paulo.
Like Camargo, many leaders of the country’s top cancer diagnosis and treatment centers closely followed the Human Genome Project. They acquired knowledge on genomics by participating in the country’s first sequencing projects, which were organized and funded by FAPESP and other institutions. In 1997, biochemists Andrew Simpson and Fernando Reinach (at the time with the Ludwig Institute for Cancer Research [LICR] and USP, respectively), and geneticist Paulo Arruda and bioinformatician João Carlos Setubal, at UNICAMP, coordinated teams from 35 São Paulo laboratories that began sequencing the genome of the bacterium Xyllela fastidiosa. It causes citrus variegated chlorosis, or yellowing, a disease that decimated the productivity of São Paulo’s orange groves.
“It was a project designed to enable these groups to perform genome sequencing, which was practically nonexistent in Brazil,” says physicist José Fernando Perez, then the Foundation’s scientific director and currently CEO of Recepta Biopharma, a biotechnology company that develops compounds to treat cancer.
Approximately three years later, the 2.7 million bases of the bacteria’s genome had been identified and ordered. The study’s results became the cover article of the July 13, 2000, issue of the journal Nature. At the time, the Human Genome Project was still in progress, and the genomes of only eight organisms—considered models in biology—had been sequenced: two viruses, one bacterium, one yeast, one worm, and one plant. The Xyllela genome was the first from an organism responsible for plant disease, and the first with commercial relevance. “It was a moment where Brazil showed that, when competing on equal terms, it could do world-class science,” says Simpson, currently scientific director of Orygen Biotecnologia, a pharmaceutical company focused on the production of antibodies, vaccines, and other medicines of biological origin.
“At that time, Brazil was one of the rare countries capable of sequencing an organism’s entire genome,” says Reinach, who left the university years ago and now runs a fund that invests in innovative companies. Since then, the genomes of nearly 19,000 organisms have been sequenced: 3,500 viruses; 14,700 bacteria; and 400 single-celled or multicellular animals and plants.

Cover stories from Pesquisa FAPESP issues nos. 50, 51, 68, and 97 (from left) dealt with projects related to genome sequencing
During the conception of the Xyllela project, oncologist Ricardo Brentani (1937–2011), then director of the Brazilian branch of LICR, decided to organize a team and participate in the sequencing. “Brentani saw Xyllela as an opportunity to bring genomics into oncology,” says Emmanuel Dias-Neto, current coordinator of the A.C. Camargo Cancer Center’s Medical Genomics Lab, which Brentani previously headed. There, as at the IEP, geneticists and other basic researchers work in collaboration with the hospital’s clinical staff to use genetic information from tumors to guide treatment and identify tumor recurrence before it becomes detectable with imaging.
In 1998, near the completion of the Xyllela genome, some laboratories already participating in the project—working together with others that had not yet entered the genomics push—used a technique developed by Dias-Neto and Simpson to sequence the internal sections of genes activated in breast, intestine, brain, throat, and other tumors, with an emphasis on those most common in the Brazilian population. Data from 280,000 sequences were deposited in a public gene databank, called GenBank, and used to assist in identifying genes in human chromosomes sequenced by the Human Genome Project groups.
Sequencing Xyllela and cancer genomes was followed in Brazil by work with other pathogens from plants (Xanthomonas citri bacteria) and humans (Leptospira sp and the Schistosoma mansoni parasite), in addition to the bovine genome. Sugarcane genes were also sequenced—enabling the production of a transgenic plant resistant to pests and herbicides—as well as those of eucalyptus. These efforts resulted in the creation of biotechnology companies such as Scylla, Alellyx, and CanaVialis. The latter two were bought by the multinational Monsanto and later closed. In Perez’s view, however, “one of the most important legacies of the genome sequencing coordinated by FAPESP was the development of bioinformatics in Brazil.”
Prior to the launch of large-scale sequencing, bioinformatics was self-taught, says UNICAMP’s João Meidanis, who majored in mathematics and opted for bioinformatics during his doctorate in the United States, when he worked on the analysis of the Escherichia coli bacterium genome. Since then, specific programs for bioinformatics have emerged at some Brazilian universities. “The community has grown, but not at the hoped-for pace, so bioinformatics remains a bottleneck for analyzing genomic data,” says Meidanis, who also runs Scylla Informática.
Arruda, from UNICAMP, assesses the era of genome sequencing as a milestone for Brazilian science. “We learned to network and manage large groups efficiently,” he says. “We also established important relationships between the university and private sector companies.”
“If we hadn’t developed these projects at the time, we might not have been ready to use this technology, which is now routine,” says USP biologist Marie-Anne van Sluys. Today she coordinates Brazil’s participation in a much more ambitious initiative: the Earth Biogenome Project, which plans to sequence the genomes of all known species of plants and animals (uni- or multicellular) over the next ten years. It will be a Herculean task. There are about 2.3 million known species, but it is estimated that in total there are 10 to 15 million.
Republish