At an industry event held in the USA in September, an American company that dominates the global DNA sequencer market announced a new machine. The latest version of its sequencer, which the company plans to start shipping next year at a cost of almost US$1 million per machine, can sequence DNA—the genetic material inside cells that holds the recipe for how living things are made and function—three times faster than its predecessors. If it performs as expected, the new model will be capable of sequencing 20,000 genomes a year at a cost of about US$200 per person, according to Wired magazine. That would represent an incredible cost reduction in just two decades. Thousands of people worked together using hundreds of machines at research centers around the world to sequence the very first draft of the human genome, which was published in 2001. It took years to determine the order of the 3 billion pairs of nitrogenous bases (the chemical units that make up DNA) in the genome, using genetic material from roughly 50 people to consolidate the equivalent of a single person’s genome. According to an estimate by the US National Human Genome Research Institute (NHGRI), which coordinated the effort, the sequencing alone cost somewhere between US$500 million and US$1 billion—the entire project cost US$3 billion.
Advances in sequencing techniques and falling costs associated with the evolution of bioinformatics tools that make it possible to analyze huge volumes of data and compare the genetic characteristics of large numbers of people are bringing genetic tests ever closer to clinical practice, even if the process is moving slower than the most enthusiastic experts predicted two decades ago, when it was first estimated that human genome sequencing would help doctors to treat, prevent, and cure diseases. Today, a variety of these tests, which might analyze parts of a specific gene or the entire genome, can be carried out at specialized centers, including in Brazil. Because the cost of sequencing the complete genome remains high—up to R$20,000 at private laboratories in Brazil—doctors more frequently request exome sequencing, used to find the cause of diseases with a high probability of having a genetic origin. The exam, which costs around R$5,000, allows alterations in multiple genes to be identified by analyzing the protein-coding sections of more than 20,000 human genes, representing less than 2% of the genome. The clinical protocols and therapeutic guidelines issued by the Brazilian Ministry of Health state that it may be offered by the country’s public healthcare system (SUS) to investigate childhood intellectual disabilities of an undetermined nature. But that does not yet happen.

Léo Ramos Chaves / Revista Pesquisa FAPESP Researcher inserts tray containing DNA into a sequencer…Léo Ramos Chaves / Revista Pesquisa FAPESP
Until a few years ago, investigations into diseases of genetic origin looked for alterations in one gene at a time. The gene suspected to be associated with a given problem was sequenced and if no mutations were found, the next gene was analyzed. “It was a slow, laborious, and expensive process,” recalls geneticist and physician Roberto Giugliani of the Federal University of Rio Grande do Sul (UFRGS). “Sequencing technologies have progressed at an exponential scale since the human genome project, expanding analysis capacity and reducing costs,” says the researcher, who also heads the rare diseases sector at Dasa Genômica, one of the largest private genetic diagnosis centers in the country.
Single-gene analyses are still carried out today, but only when it is already known which genetic alteration to look for, such as with some rare diseases that despite individually affecting a very small percentage of the population, altogether affect 6% of the world’s population and some 13 million Brazilians. They are most often caused by defects in a single gene and are thus known as monogenic diseases. More comprehensive tests that include thousands of genes or millions of points on the genome (or even complete sequencing), which would be of interest to a much larger number of people, have been adopted in the diagnosis of many rare diseases. They are becoming more prominent in the portfolios of private companies and laboratories, which in recent years have also started to sell tests directly to consumers for just a few hundred reais.
Dubbed recreational tests, they do not require a medical prescription and are used to compare sections of an individual’s genetic material with data stored in international databases. Some, known as ancestry tests, provide information about the probable geographical origin of a person’s ancestors. Others aim to determine the most suitable diet or type of physical activity for an individual based on their genetic information, or even to check whether they are predisposed to certain diseases that affect a high proportion of the population, such as type 2 diabetes, heart disease, or cancer. “These direct-to-consumer genetic tests have become the greatest genetic product for the general population,” reports geneticist Michel Naslavsky, a professor at the University of São Paulo (USP) and researcher at the Albert Einstein Education and Research Center who studies gene alterations that affect disease risk.
The amount and type of genetic information analyzed by direct-to-consumer tests, however, do not accurately diagnose or estimate the risk of these diseases occurring or provide specific guidance on how to prevent them. Like monogenic diseases, most common illnesses also have a genetic component. But there is a fundamental difference. In common diseases this aspect depends on the interaction between hundreds or thousands of genes (because they are polygenic), each contributing slightly to the final outcome. Changes therefore have to occur in a large number of these genes for the disease to manifest. Finding a fault with one or two of them usually means nothing—the same is true for complex traits such as muscle strength, body fat, or height—since environmental factors also play a part.
“Tests sold directly to the consumer use genetic data that can tell them some interesting facts about themselves, such as their ancestry, but they cannot provide evidence for medical use,” says geneticist and physician Rayana Maia, a professor at the Federal University of Paraíba (UFPB) and director of the Ethics Department of the Brazilian Society of Medical Genetics and Genomics (SBGM). “Many of the genetic variants [altered versions of genes] identified in these tests are not associated with a major risk of disease. The result can lead more to anguish than any benefit,” she concludes.
Léo Ramos Chaves / Revista Pesquisa FAPESP … and then analyzes the resultLéo Ramos Chaves / Revista Pesquisa FAPESP
Whether for clinical or recreational use, the fact is that genetic tests represent a billion-dollar market, that is growing at a rate of almost 10% per year, according to some analyses. In a recent report, American market analytics company Straits Research estimated that the genetic testing market was worth US$14.8 billion worldwide in 2021 and is forecast to reach US$36 billion by 2030.
Faced with so much potential, a number of questions remain: who are genetic tests aimed at? And when should they be used?
The answer is: It depends. If someone wants to know a little bit about where their ancestors came from, they can find out whenever they want. But it should be kept in mind that the result may vary depending on the company offering the test. The person’s genetic data is compared against information from tens of thousands of individuals stored in the test provider’s database. In general, these databases comprise data from people living in Europe and the USA with mostly Caucasian ancestry. “A test like this carried out with the DNA of a Brazilian, for example, may not identify the existence of ancestors native to South America or some regions of Africa,” says Maia.
If the aim is to determine the risk of developing a disease in the future or transmitting a genetic alteration associated with a serious disease to a child, the answer is less simple. “Genetic testing for diagnoses should only be conducted with medical guidance and genetic counseling, because if there is no specific reason for doing it, the result can do the individual more harm than good,” says geneticist José Cláudio Casali da Rocha, founder and coordinator of the Oncogenetics Center at the A.C.Camargo Cancer Center in São Paulo. “A person could live their whole life in fear of developing a disease that may never manifest,” he explains.
Doing a test, even a recreational one, to find out about your genetic profile and whether your cells contain gene variants that may be somehow associated with diseases can be problematic. “Genetic tests are powerful diagnostic tools that often make it possible to end a long search for the origin of a disease of unknown cause,” says USP endocrinologist Alexander Jorge, who specializes in genetic diseases. “But when performed for no specific reason, without a question to be answered, they can reveal alterations that have no clinical significance, which can lead to misinterpretations.”
All humans are 99.9% identical on a genetic level. This means that the sequence of four nitrogenous bases—adenine (A), cytosine (C), guanine (G), and thymine (T)—in the DNA of any two people, whether they are identical twins or were born on opposite sides of the planet, is almost exactly the same. What makes one human being different from another is the 0.1%, which represents 3 million pairs of these bases. “The number of variations in the genome of two healthy human beings is huge, but very few have a known influence on health and it is often difficult to identify which of them are associated with a problem,” explains Giugliani, from UFRGS.
Progress in sequencing techniques and gene analysis over the last few decades has dramatically increased the number of genes and variants known to play a role in disease. The clearest and most significant advance has been made in the field of monogenic diseases, where the alteration of a single gene has a devastating impact, enough to significantly impair normal functioning. These illnesses are usually serious—some fatal, such as spinal muscular atrophy—and begin to manifest in childhood.
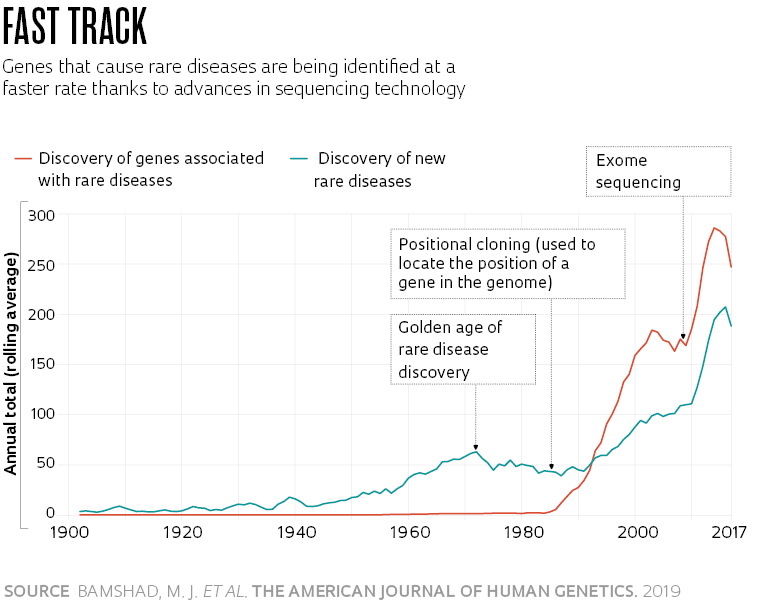
In a comment published in The American Journal of Human Genetics in 2019, pediatrician Michael Bamshad of the University of Washington, USA, and his colleagues reported that the description of these diseases and identification of the genes that cause them has progressed rapidly since the mid-1980s, alongside advances in DNA sequencing and the Human Genome Project (see graph). Until then, only about 40 genes had been characterized as causing some of these diseases. By November 21, alterations in 4,721 genes associated with 7,286 rare diseases had been identified, according to the Online Mendelian Inheritance in Man (OMIM) database.
In these cases, genetic tests play an important role in improving diagnosis and allowing for early treatment when available, in addition to prevention through genetic counseling for people found to have the altered version of a gene that plan to have children. “Some rare diseases are very difficult to recognize based on the clinical situation alone,” says USP’s Alexander Jorge. “In these cases, exome sequencing shortens the time taken to make a diagnosis, reduces costs, and allows medical professionals to advise the family.”
Identifying the cause of a genetic disease improves quality of life by allowing doctors to choose the most effective treatments for the symptoms and avoid medications that can make them worse. It also helps to prepare parents and caregivers for how the disease might progress. “There is not yet any treatment that can alter the genes and cure these diseases, but for many of them, we can treat the symptoms and offer the child a better quality of life,” says geneticist Maria Rita Passos-Bueno, who runs the Genetic Test Laboratory (LabTEG) at USP’s Center for Human Genome and Stem Cell Studies (CEGH-CEL) together with geneticist Mayana Zatz.
One of the fields that has benefited most from genetic testing is oncology. Although different types of cancer can manifest in different ways (fast or slow progression; high or almost zero potential for a cure, etc.), they are essentially genetic diseases. Alterations in one or more genes modify cell programming and cause them to multiply uncontrollably, damaging the organs.
In recent years, oncologists have begun using a range of tests to track genome changes that are known to increase the likelihood of cancer in an effort to reduce the risk of developing the disease. They have also begun using genetic information from tumors to select the most suitable drugs and doses for each patient, as well as treatments to be avoided. International consortiums and regulatory agencies such as the US Food and Drug Administration (FDA) have already identified more than 250 drugs (several for oncological use) that should be prescribed based on the DNA profile of the patient. “Genetic tests help determine which biochemical pathways the tumor is using to grow and what drugs could inhibit or block its function,” says Casali, from A.C.Camargo.
One challenge is to find a way of extracting information from the genetic profile that can be used to estimate the risk of common diseases
While studying the genetic profile of 192 Brazilian women with a type of breast cancer whose growth is stimulated by the female hormone estrogen, Casali and his colleagues found that not all patients responded as expected to treatment with tamoxifen, a compound that inhibits the production of molecules on the surface of tumor cells to which the hormone binds, reducing its impact. An inert version is administered, which is then activated in the liver by the enzyme CYP2D6. Women with an altered CYP2D6 gene produce a less functional version of this enzyme. According to the results of the study, published in Clinical and Translational Science in 2020, this characteristic partly explains why there was less of the drug in the blood of these individuals than is needed to prevent the cancer from returning.
In a phase 2 clinical trial, a team led by oncologist Fabrice André of the Gustave Roussy cancer center in Paris provided two different treatments to 238 breast cancer patients with metastasis: 157 received specific medication to combat the effects of the genetic alterations characteristic of their tumors and 81 underwent traditional chemotherapy. After almost two years of monitoring, the disease’s progression had been contained for an average of nine months in the first group and for less than three months in the second, according to an article published in Nature in July.
One remaining challenge is to find ways of extracting reliable information from a person’s genetic profile that can be used to estimate the risk of developing common diseases that affect much of the population, such as heart disease or diabetes. According to genetic studies, these diseases are usually caused by alterations in a large number of genes. Every alteration contributes a little to the disease risk—the most expressive variants account for just 0.2% of the chance of developing the disease—but their impacts add up.
In recent years, geneticists have begun testing the concept of using a mathematical formula (known as the polygenic risk score) to estimate the probability of developing these diseases. To put it simply, this score or risk profile represents the sum of all of an individual’s risk-associated genetic variants, weighted by the extent to which each alteration contributes. According to a review article published in Nature Reviews Genetics in 2018 by Ali Torkamani, a chemist from Scripps Research, USA, who specializes in informatics and genomics, recent studies are starting to demonstrate the usefulness of these scores for identifying groups of individuals more susceptible to developing certain diseases and who could benefit from medical monitoring or interventions.

Léo Ramos Chaves / Revista Pesquisa FAPESP Some genetic tests are less accurate for mixed-race populations, such as BraziliansLéo Ramos Chaves / Revista Pesquisa FAPESP
Three years ago, a group led by cardiologist and geneticist Sekar Kathiresan at Massachusetts General Hospital’s Center for Genomic Medicine, USA, developed a polygenic risk score for obesity based on information from 2.1 million gene variants with a known influence on body mass index (BMI). The tool was tested using DNA and health data from 300,000 people. The score quantified the risk of becoming severely obese (meaning a BMI of more than 40—a BMI of between 18 and 25 is considered healthy) and the consequent increased likelihood of developing certain diseases. The 10% of individuals with the highest number of risk-associated variants were 25 times more likely to become severely obese than the 10% with the lowest number of these alterations. Among the former, the risk of developing diabetes, a thromboembolism, or hypertension was 70%, 40%, and 35% higher respectively than in the latter group, according to the results, published in the journal Cell in 2019.
Despite their promise, polygenic risk scores are still a long way from being used in a clinical setting. They first need to be validated with large samples of populations from different parts of the planet. It is already known, for example, that having been created based on genetic data from groups of mostly European ancestry, these tools are less accurate for more diverse populations. Brazilian geneticist Bárbara Bitarello, currently on a postdoctoral fellowship at the University of Pennsylvania, USA, found that a score designed to predict people’s height—a characteristic that is easy to measure—was four to five times less accurate for groups with African ancestry, according to a 2020 paper in Genes, Genomics, Genetics.
In these situations, ancestry is not merely a curiosity, it is medically significant. “Research is beginning to show that in order to calculate the genetic risk of developing a health problem, it will be important to take ancestry into account,” says geneticist Eduardo Tarazona Santos of the Federal University of Minas Gerais (UFMG), who studies the ancestry of Brazilians. He explains that gene variants can have different effects depending on whether they were inherited from African, European, or Amerindian populations.
Tarazona participated in a study led by USP’s Michel Naslavsky and Claudia Suemoto, the results of which were published in Molecular Psychiatry in September, which revealed that a gene variant that increases the risk of developing Alzheimer’s and another that protects against the disease have less of an influence in populations of African origin than in descendants of European ancestry. “For now, the results of polygenic risk scores need to be treated with caution. More than 2,000 such indices have already been proposed, but only a few dozen have been tested on Latin American populations,” says Tarazona.
Projects
1. CEGH-CEL – Human Genome and Stem Cell Research Center (no. 13/08028-1); Grant Mechanism Research, Innovation, and Dissemination Center (RIDC); Principal Investigator Mayana Zatz (USP); Investment R$54,769,696.92.
2. INCT 2014 – Aging and genetic diseases: genomics and metagenomics (no. 14/50931-3); Grant Mechanism Thematic Project; Principal Investigator Mayana Zatz (USP); Investment R$2,913,294.82.
Scientific articles
BAMSHAD, M. J. et al. Mendelian gene discovery: Fast and furious with no end in sight. The American Journal of Human Genetics. Vol. 105, pp. 448–55. Sept. 5, 2019.
NARDIN, J. M. et al. The influences of adherence to tamoxifen and CYP2D6 pharmacogenetics on plasma concentrations of the active metabolite (z)-endoxifen in breast cancer. Clinical and Translational Science. Vol. 13 pp. 284–92. 2020.
ANDRÉ, F. et al. Genomics to select treatment for patients with metastatic breast cancer. Nature. July 3, 2022.
TORKAMANI, A. et al. The personal and clinical utility of polygenic risk scores. Nature Reviews Genetics. Vol. 19, pp. 581–90. Sept. 2018.
KHERA, A. V. et al. Polygenic prediction of weight and obesity trajectories from birth to adulthood. Cell. Vol. 177, pp. 587–96. Apr. 18, 2019.
BITARELLO, B. D. & MATHIESON, I. Polygenic scores for height in admixed populations. Genes, Genomes, Genetics. Nov. 2020.
NASLAVSKY, M. S. et al. Global and local ancestry modulate APOE association with Alzheimer’s neuropathology and cognitive outcomes in an admixed sample. Molecular Psychiatry. Sept. 7, 2022.