The air around us is not an inert space containing nothing but oxygen, nitrogen, water vapor, dust, and pollution. It is a living environment in which millions of microorganisms are continuously born, reproduce, and then die. In 2019, analyses of air samples from Singapore and the Brazilian Amazon indicated that bacteria predominate during the day and fungi at night, with little variation throughout the year. Now, new research in the Asian city-state has found that this division is maintained at up to a thousand meters (m) altitude. Above this limit, bacteria adapted to high levels of solar radiation, rarer at lower levels, reign.
This study is an example of how field research in biology increasingly relies on mountains of data. The Singapore group, led by German chemist Stephan Schuster from Nanyang Technological University (NTU), examined 795 air samples collected at varying altitudes, identifying roughly 10,000 species of microorganisms and most impressively, processed 2.2 terabytes of data, equivalent to 2 million megabytes.
Biologists carrying out field research are facing the kind of major changes that geneticists experienced 20 years ago, when the number of sequenced genomes grew exponentially, leading to new working methods. Over the last 15 years, geneticists and ecologists have started using more powerful DNA sequencers capable of deciphering multiple genomes at the same time, which have allowed for extensive surveys of organisms living in the soil, air, water, and even inside other beings.
Now, other devices that have become more widely used in the last five years, such as drones and recorders designed for field work, are also storing and processing previously inconceivable amounts of data and allowing scientists to make better use of their time during expeditions in forests, plains, and savannas. While these new technologies have made the work of researchers easier, they also demand a more in-depth knowledge of mathematics, statistics, and programming. Working with specialists from other fields is now imperative.
“Analyzing the microbiome in the air was only possible thanks to collaborations between biologists, climatologists, programmers, and the engineers who adapted the instruments for the research plane used to collect data for this study,” highlights Ana Carolina Martins Junqueira, a biologist from the Federal University of Rio de Janeiro (UFRJ) who participated in the study in Singapore.
In the early 2000s, while studying her master’s and PhD at the University of Campinas (UNICAMP), she sequenced the mitochondrial DNA of blow flies—one genome at a time. In 2011, when she arrived for a postdoctoral fellowship at Pennsylvania State University, USA, she found more powerful machines: next-generation sequencers. With these, she was able to simultaneously sequence the genomes of all the organisms living in the viscera of two fly species, Chrysomya megacephala and Musca domestica, collected in Brazil and the USA. She found 431 species of bacteria, including Helicobacter pylori, which causes ulcers and stomach cancer, as described in an article she published in Scientific Reports in November 2017.
In 2014, Junqueira accompanied Schuster, her supervisor in the USA, on a trip to NTU’s Singapore Centre for Environmental Life Sciences Engineering (SCELSE), where she participated in the intensive search for communities of microorganisms. The first results revealed the daily cycle of microorganisms that live in the air near ground level.
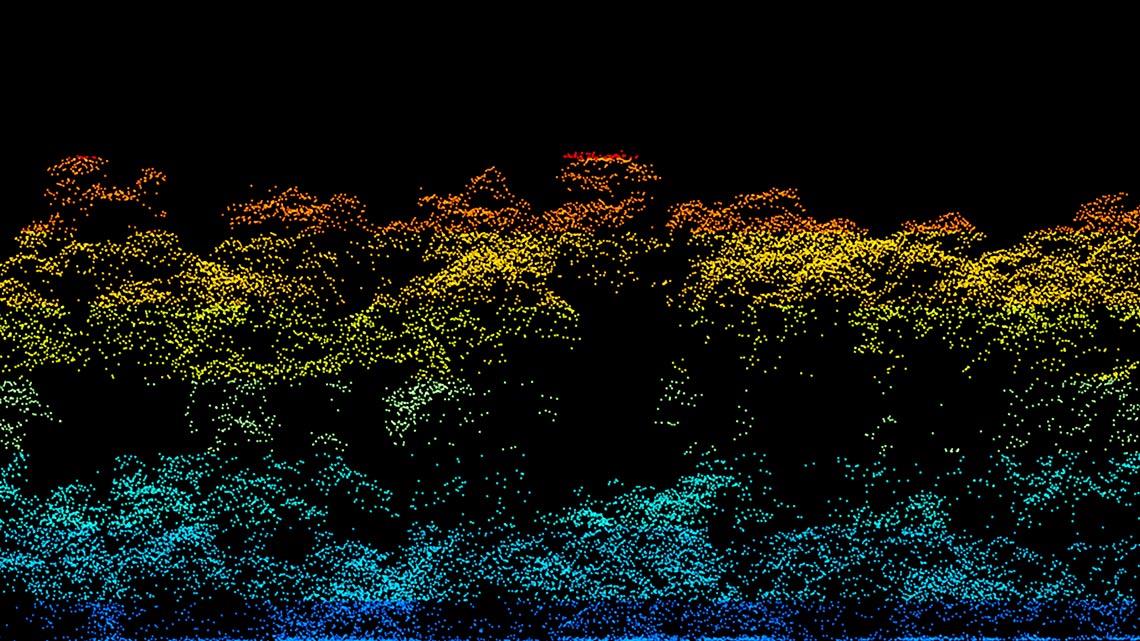
A complementary study that used other benchmarks—samples collected near the ground, from a meteorological tower, and from a research plane at altitudes of up to 3,500 m—indicated that the daily cycle breaks down at an altitude of approximately 1,000 m and that there is a stratification: there are more fungi closer to the ground, more bacteria at intermediate altitudes, and a lower diversity of microorganisms above 1,000 m, where there is an abundance of radiation-tolerant bacteria.
In parallel, scientists from Paraná and Amazonas collected 16 air samples at altitudes of 2 m and 26 m from the Amazon Tall Tower Observatory (ATTO), located in an area of rainforest area 150 kilometers (km) from Manaus, in August 2018 and March 2019. “Little was known about the types of microorganisms circulating in the air in the Brazilian rainforest,” says Luciano Huergo, a biologist from the Federal University of Paraná (UFPR), Matinhos campus, who led the study. The bacteria of the genera Beijerinckiaceae and Azospirillum were abundant and drew attention for its ability to transform atmospheric nitrogen (N2) into ammonium (NH4), which plants can absorb. This means they could potentially function as a natural fertilizer.
In 2009, while studying his master’s degree with the Biological Dynamics of Forest Fragments Project (PDBFF) at the Brazilian National Institute of Amazonian Research (INPA) in Manaus, biologist Marconi Campos-Cerqueira was overwhelmed by the volumes of data he collected. He had to identify the birds in the forest just by listening to them, without seeing them. Recording equipment made the task easier, but another problem soon arose: he recorded so many hours of sound that there was no way he could listen to it all himself.

Research plane used to collect air samples in SingaporeScelse/NTU
Luckily, he met American biologist Mitchell Aide, who was developing a computer program to identify bird species from their singing and invited him to work with him at the University of Puerto Rico. Cerqueira accepted and did his PhD with Aide, helping create the Automated Remote Biodiversity Monitoring Network (ARBIMON), which separates and identifies the sounds of each species. The latest version, described in the journal Ecological Informatics in September 2020, was 89% accurate at automatically identifying 24 species of birds and frogs in Puerto Rico.
“Acoustic monitoring can be used in combination with other methods, because we still need to go out into the field to learn about the environment, habits, and diet of each species,” says Cerqueira, who has been chief scientist at the nongovernmental organization Rainforest Connection since 2017. Freely available to researchers, the computer platform stores 2,163 research projects, 57 million recordings, 29,000 analyses, and 2,000 sounds of birds, amphibians, and other terrestrial and marine animals from South America, Asia, Africa, and Europe.
Cerqueira’s colleague and ARBIMON user Alexandre Camargo Martensen, a biologist from the Federal University of São Carlos (UFSCar), Buri campus, is studying 120 areas near the Atlantic Forest. His team has recorded almost one million minutes of bird, amphibian, and mammal sounds.
Based on these recordings and conversations with local residents, the UFSCar team found 15 specimens of an orange and lime green frog called Phrynomedusa appendiculata in an area of Atlantic Forest in Capão Bonito, São Paulo State. The species has not been seen since 1970, as reported in Zootax in January.
Like Martensen, biologist Alexandre Uezu from the Institute for Ecological Research (IPÊ) in Nazaré Paulista spent most of his master’s and PhD doing fieldwork, but now spends most of his time in front of a computer, organizing, identifying, and analyzing sounds and images. He monitors changes in an area of Atlantic Forest in Cantareira State Park, in Greater São Paulo, another in Pontal do Paranapanema in the west of the state with funding from China Three Gorges (CTG Brasil), and a third in Alto Paranapanema, in the south.
Devices installed at 500 sampling points have already recorded around 1.5 million minutes of sound. He also collects images via satellite and more recently by lidar (light detection and ranging), which records variations in light reflected by trees, used by PDBFF to survey the Amazon rainforest since 2013 (see Pesquisa FAPESP issue no. 205). In this case, the laser beams determine the height of the trees and when they are able to reach the ground, they also detail the relief.
To calculate the expected growth of a restored forest in an area connecting state and federal conservation units, Uezu and his team used lidar data from manned flights made in 2015, 2016, and 2017. They identified a 35% growth in forest biomass between the two reserves.
More zoom, less stress
Fabiano Rodrigues de Melo, a biologist from the Federal University of Viçosa (UFV), first heard about drones in 2014, when they were being used to find koalas in Australian forests. In 2017, for a study funded by the Boticário Group Foundation, he ordered his first drone for a company based in Rio de Janeiro that also made a computer program capable of counting northern muriquis (Brachyteles hypoxanthus), thus removing the need to personally watch hours upon hours of footage.
His current drone, purchased with funding from the Wildlife Conservation Society, is small enough to fit in his backpack. With its powerful zoom, he can film animals from 30 m away without scaring them. It also makes it easier to identify animals by differences in their facial pigmentation. “Before, I had to get to within 15 m and the muriquis would usually run away—maybe they thought the drone was a predator,” he says. “In dense forests, I can only see animals at the tops of the trees, but in secondary forests I’ve seen tamarins, which occupy lower areas, and even deer, capybaras, tayras, coatis, anteaters, and dozens of bird species on the ground.”
Devices that identify animals and plants by sound, image, or DNA facilitate the life of researchers and contribute to environmental impact studies required by government licensing agencies or carbon sequestration projects. In order to be effective, however, the information they generate must be carefully managed. “The avalanche of data changes how they have to be analyzed and stored,” stresses UFSCar biologist Alexandre Camargo Martensen.
According to him, the collected data and the research plan need a script that recognizes the successive versions and is stored in appropriate databases while the research is carried out. “We write these analysis scripts ourselves in R or Python,” he says.
“Since data are so much more easily obtained now, we have to pay extra attention to how they are stored and analyzed,” adds Alexandre Uezu, from IPÊ. A false negative result—failing to identify a species that exists—is better than a false positive, he says. “You mustn’t get into a frenzy of collecting data without first clearly defining the objective of the study,” recommends Marconi Campos-Cerqueira. “It is important to define the guiding hypotheses before starting to collect the data.”
Projects
1. Forest transition governance in the Atlantic Forest: Increasing our knowledge of forest restoration and ecosystem services (nº 18/20501-8); Grant Mechanism Biota Program; Principal Investigator Alexandre Camargo Martensen (UFSCar); Investment R$388,961.59.
2. Resilience to climate change in multifunctional landscapes (nº 19/19429-3); Grant Mechanism Biota Program; Principal Investigator Alexandre Uezu (IPÊ); Investment R$121,162.26.
Scientific articles
CLARE, E. L. et al. Measuring biodiversity from DNA in the air. Current Biology. v. 32, n. 3, p. 693–700. feb. 7, 2022.
COSTA, D. P. da. et al. Forest-to-pasture conversion modifies the soil bacterial community in Brazilian dry forest Caatinga. Science of Total Environment. v. 41, 151943. mar. 1, 2022.
DRAUTZ-MOSES, D. I. et al. Vertical stratification of the air microbiome in the lower troposphere. PNAS. v. 119, p. e2117293119. feb. 15, 2022.
GUSAREVA, E. S. et al. Microbial communities in the tropical air ecosystem follow a precise diel cycle. PNAS. v. 116, no. 46, p. 23299–308. nov. 12, 2019.
JUNQUEIRA, A. C. M. et al. The microbiomes of blowflies and houseflies as bacterial transmission reservoirs. Scientific Reports. v. 7, n. 1, 16324. nov. 24, 2017.
LEBIEN, J. et al. A pipeline for identification of bird and frog species in tropical soundscape recordings using a convolutional neural network. Ecological Informatics. v. 59, 101113. sept. 2020.
LYNGGAARD, C. et al. Airborne environmental DNA for terrestrial vertebrate community monitoring. Current Biology. v. 32, n. 3, p. 701–7. feb. 7, 2022.
MORAES, L. J. C. L. et al. Rediscovery of the rare Phrynomedusa appendiculata (Lutz, 1925) (Anura: Phyllomedusidae) from the Atlantic Forest of southeastern Brazil. Zootaxa. v. 5087, p. 522–40, 2022.
NASCIMENTO, L. A. do et al. Acoustic metrics predict habitat type and vegetation structure in the Amazon. Ecological Indicators. v. 117, 106679. oct. 2020.
SOUZA, F. C. S. et al. Influence of seasonality on the aerosol microbiome of the Amazon rainforest. Science of The Total Environment. v. 760. mar. 15, 2021.