When the Zika virus caught Brazil by surprise at the end of 2015, it had likely already been circulating throughout the country for just over two years. It is relatively silent, because despite causing some fever, body and joint pain, marks on the body, and other symptoms that previously escaped diagnosis because of their similarity to dengue and Chikungunya, these are nothing in comparison with the dramatic birth of children with problems affecting brain development, namely fetal microcephaly. Now some faster, more reliable and practical means of diagnosing the virus are available, which can allow earlier treatment and public health measures. And as for the virus’s ability to destroy neurons, how about using it for good?
This is the idea of a group led by geneticist Mayana Zatz of the Institute of Biosciences at the University of São Paulo (IB-USP) and coordinator at the Center for Research on the Human Genome and Stem Cells (HUG-CELL), one of the Research, Innovation, and Dissemination Centers (RIDC) supported by FAPESP. In an article published at the end of April in Cancer Research, the researchers showed that in a mouse model, the Zika virus can be used as a tool to treat aggressive tumors of the human central nervous system. This is the first time this has been done in a live model.
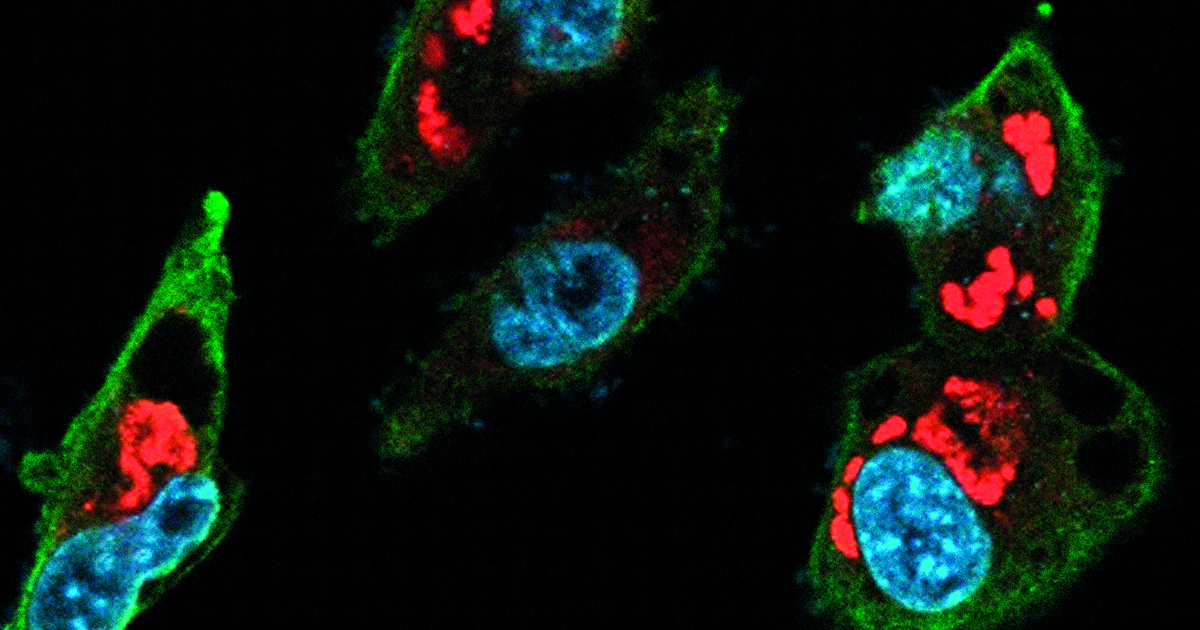
Zatz led the research alongside Oswaldo Keith Okamoto, also of IB-USP and HUG-CELL. “Our studies and those done by other groups showed that the Zika virus causes microcephaly because it infects and destroys the neural stem cells in the fetus, preventing new neurons from being formed. This is when we had the idea to investigate whether it also would affect tumor stem cells in the central nervous system,” says Okamoto.
In vitro experiments began with three human tumor strains that cause cancer in the central nervous system. The researchers added increasing quantities of virus to the tumor cells in culture and monitored them using immunofluorescence microscopy. “We noticed that small quantities of the Zika virus were sufficient to infect the cells from the central nervous system tumors. Prostate cancer [cells] were infected, but in a much smaller proportion,” he reports. When they compared the effect in healthy neural stem cells obtained from induced pluripotent stem cells (IPS, adult cells reprogrammed in the laboratory to behave like stem cells) with the tumor cells, they found that the virus more efficiently destroys these tumor cells. “We also exposed mature neurons to the virus and saw that they were not infected or destroyed by the pathogen,” adds Okamoto. “This is great news, since our goal is to specifically destroy tumor cells,” adds Mayana Zatz.
The tests then progressed to mice with human tumors in their brains; some of these animals were injected with a small dose of Zika, and in this treatment group there was a significant reduction in tumor volume. The group intends to move on to clinical trials. “The idea is to start with two or three patients who have not responded to conventional treatments, and if the strategy works, extend it to a larger group,” states Zatz. The group has filed a patent for the therapeutic protocol adopted in rodents.
Machine learning
The Zika virus was also the starting point to develop a platform capable of diagnosing various diseases via metabolic markers in the blood of patients. “In this case, the diagnostic accuracy exceeds 95%,” says Rodrigo Ramos Catharino, a pharmacologist at the School of Pharmaceutical Sciences at the University of Campinas (UNICAMP). “One of the major advantages is that the method does not lose sensitivity, even if the virus mutates.”
The method combines mass spectrometry technology, which permits the identification of tens of thousands of molecules present in blood plasma using an artificial intelligence algorithm that can spot patterns associated with illnesses, whether viral in origin like bacterial diseases, fungal, or even genetic. The results, which were part of doctoral research by pharmacologist Carlos Melo and were published in April in the journal Frontiers in Bioengineering and Biotechnology, reveal the ability to detect the virus even 30 days after the beginning of the infection. “No available diagnostic kit has the sensitivity to detect Zika infection after the end of the acute phase. For example, our method could be useful to analyze bags of blood for transfusion,” says Catharino.
In the field
While the Zika virus can be recognized and even useful in the laboratory, in the outside world it is still difficult to detect with the swiftness needed to minimize its harmful effects.
Promising results for use in the field come from Iranian-American geneticist Pardis Sabeti’s group at the Broad Institute in the United States, as published in April in Science. This work was led by virologist Catherine Freije, a doctoral student at Harvard University, and systems biologist Cameron Myhrvold, a postdoctoral fellow at the Broad Institute; it helped refine the SHERLOCK diagnostic platform (named for Specific High-sensitivity Enzymatic Reporter unLOCKing) developed by another group at the same institute. The modified method was called HUDSON (Heating Unextracted Diagnostic Samples to Obliterate Nucleases) and permits the detection of RNA and DNA through an enzymatic reaction that can even take place on paper strips, far from the laboratory. This is possible thanks to chemical and thermal treatment to inactive enzymes that otherwise degrade genetic targets.
The SHERLOCK-HUDSON methods recognize viruses in the sample (blood, saliva, or urine) using the CRISPr-Cas13 system, which can find extremely specific sequences in the viral genome, only showing a match when there is a high concordance between the target and the model transported by the system. “This system is unique in the CRISPr family, because when a sequence is recognized, the Cas13 enzyme can break an additional RNA sequence,” Freije and Myhrvold described in a joint email. “This is particularly useful in the diagnostic context, because we can introduce reporter molecules that create a fluorescent or visual mark when they are broken.”
In under two hours, this method detected the viruses that cause Zika and dengue fever in samples from Brazilian patients collected between 2015 and 2016 within a project led by virologist Mauricio Lacerda Nogueira of the São José do Rio Preto School of Medicine (FAMERP). “The dengue and Zika viruses are very similar, and often produce conflicting results in the tests. This SHERLOCK platform diagnosed even the samples that were positive for more than one virus with 100% accuracy,” said Nogueira.
He adds that this type of technology permits the test to be tailored specifically. “If an epidemic of a new virus breaks out, the kit can be quickly developed with the reagents and brought to the site. However, we are still a few years away from a commercial application.” The detailed recognition of genetic sequences can allow specific mutations to be identified, such as in proteins associated with the development of fetal microcephaly, or can allow researchers to track the geographical origin of the lineage present in a patient. The method is in the refinement phase, and may complement other techniques which have already been developed, such as sequencing in the ZiBRA project (see Pesquisa FAPESP issue no. 256).
With collaboration by Maria Guimarães
Projects
1. The Human Genome and Stem Cell Research Center (HUG-CELL) (no. 13/08028-1); Grant Mechanism Research, Innovation, and Dissemination Centers (RIDC); Principal Investigator Mayana Zatz (USP); Investment R$27,308,809.64 (for the entire project).
2. Epidemiological study of dengue (serotypes 1 to 4) in prospective cohort from São José do Rio Preto, São Paulo, Brazil, from 2014 to 2018 (no. 13/21719-3); Grant Mechanism Thematic Project; Principal Investigator Maurício Lacerda Nogueira (Famerp); Investment R$2,893,092.30.
3. Metabolomics and lipidomics of the Zika virus, from the mosquito to the patient (no. 16/17066-2); Grant Mechanism PhD Grant; Principal Investigator Rodrigo Ramos Catharino (UNICAMP); Scholarship Beneficiary Carlos Fernando Odir Rodrigues Melo Investment R$108,420.39.
Scientific articles
KAID, C. et al. Zika virus selectively kills aggressive human embryonal CNS tumor cells in vitro and in vivo. Cancer Research. Online. Apr. 26, 2018.
MYHRVOLD, C. et al. Field-deployable viral diagnostics using CRISPR-Cas13. Science. Vol. 360, no. 6387, pp. 444–8. Apr. 27, 2018.
MELO, C. F. O. R. et al. A machine learning application based in random forest for integrating mass spectrometry-based metabolomic data: A simple screening method for patients with Zika virus. Frontiers in Bioengineering and Biotechnology. Online. Apr. 11, 2018.