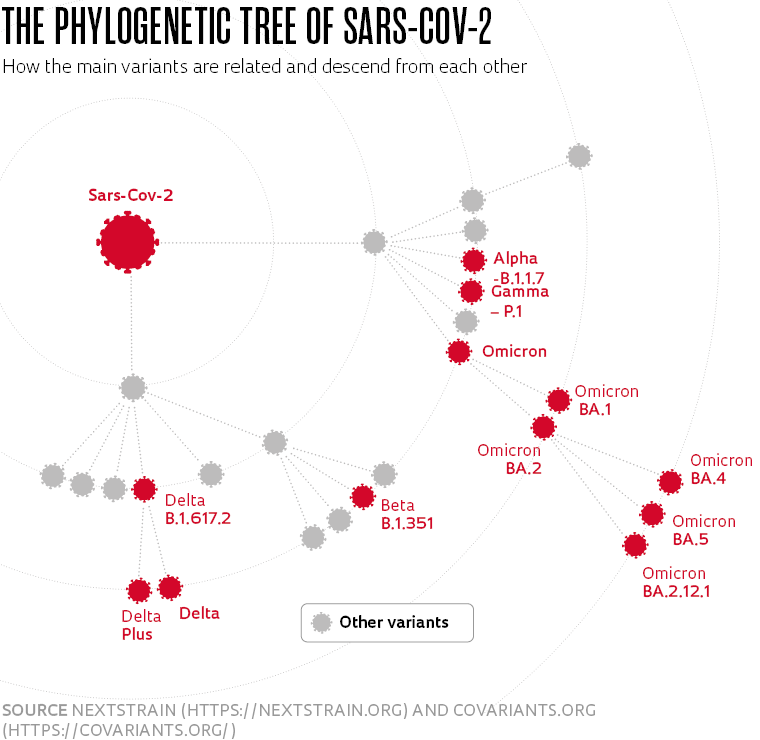
What variants of the virus that causes COVID-19 are circulating in Brazil at the moment and what is their impact on the health of the population? Do new strains of SARS-CoV-2 spread more easily? Do they cause more or less serious forms of the disease? Are vaccines and tests still effective? These are some of the questions being asked by genome surveillance projects monitoring which mutations are present in the virus as it circulates around Brazil. It is specialists in this area who raise the alarm if a new variant of concern—a strain that potentially poses a serious risk to public health, either because it is more transmissible or more lethal—emerges.
According to the international platform GISAID, only 0.47% of the 31 million cases of the disease recorded in Brazil have been sequenced to decode their genetic makeup, well below the 5% recommended by experts. In absolute numbers, fewer than 150,000 genomes were analyzed. The percentage was even lower in the first year of the pandemic due to a lack of funding, equipment, and supplies. GISAID is the leading international database for sharing data on the genetic code of the novel coronavirus and any mutations detected.
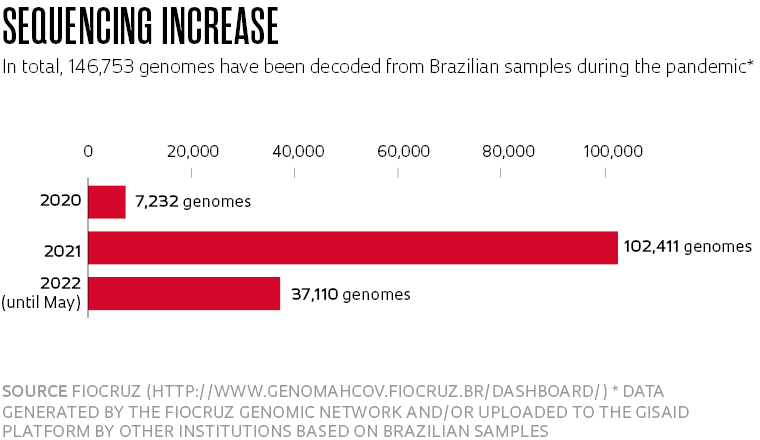
Although genome surveillance in the country has been improving since late 2021 (see infographic), scientists and administrators are still searching for funding to ensure their work can be continued—or expanded. The good news is that the two major Brazilian genome surveillance networks have recently received support from the UK Health Safety Agency (UKHSA) to intensify their work on genetic sequencing and identify possible new variants in the country.
Researchers from the FIOCRUZ Genomic Network, funded by Brazil’s Ministry of Health (MS) and coordinated by scientists from the Oswaldo Cruz Foundation (FIOCRUZ), and the National COVID-19 Omics Network (Rede Corona-ômica BR), funded by the Brazilian Ministry of Science, Technology, and Innovation (MCTI), regularly participate in working meetings with members of the New Variant Assessment Platform (NVAP), which was created by the British government last year to reinforce infrastructure around the world and help find new variants. Chile, Singapore, Ethiopia, Kenya, Nigeria, Pakistan, and Trinidad and Tobago are also involved in the initiative.
“The British government agency is not our only source of funding, but it is important to allowing us to continue with our analyses and generate genomic information,” stresses Marilda Siqueira, head of the Respiratory Virus and Measles Laboratory at the Oswaldo Cruz Institute (IOC/FIOCRUZ). The research, development, and education center serves as a COVID-19 reference point for the MS in Brazil and for the World Health Organization (WHO) in the Americas.
Siqueira, head of the FIOCRUZ Genomic Network, says that the British government’s support could enable more FIOCRUZ units to work on analyzing the SARS-CoV-2 genome. Of the institution’s 11 centers, only eight currently carry out this task: in the states of Amazonas, Ceará, Pernambuco, Piauí, Bahia, Rio de Janeiro, Minas Gerais, and Paraná.
“Genomic analysis is an expensive and complex activity. It requires not only qualified personnel, but also expensive equipment and supplies,” explains the scientist. According to Siqueira, the British funding is estimated to last for one year, and will allow the FIOCRUZ network to purchase inputs and train personnel in bioinformatics, data analysis, and epidemiological surveillance. “We started receiving inputs in April and we have already begun some training.”
On another front, the National COVID-19 Omics Network reached an agreement with the Brits a little later and inputs had not yet arrived at the end of June, although experts from the UK were already holding workshops with the team. Formed in 2020, the network involves 19 research centers, mainly based at universities. “The British are seeking to support established networks and provide inputs so that we can improve our genomic sequencing and surveillance efforts,” says virologist Fernando Spilki, head of the network and a professor at Feevale University in Nova Hamburgo.
“For us, the most important thing in relation to this project is the training of personnel. The UK has great expertise in crossing genomic surveillance data with epidemiological information,” says the researcher. “It may seem very simple to correlate a particular variant or sublineage with an outbreak or rise in cases, but many aspects need to be considered, such as social, geographic, economic, and temporal issues.”
One of the COVID-19 Omics Network’s projects supported by NVAP is led by Ester Sabino, a physician and professor at the Institute of Tropical Medicine (IMT) of the University of São Paulo’s School of Medicine (FM-USP). Sabino is head of the UK-Brazil Joint Center for Arbovirus Discovery, Diagnosis, Genomics, and Epidemiology (CADDE), funded by FAPESP and the Medical Research Council, a British funding agency (see Pesquisa FAPESP issue nº 304).
In 2021, she started a genome surveillance project with investment from retail company Magalu, as part of an initiative run by Grupo Mulheres do Brasil (“Brazilian women’s group”). The initial aim was to set up a group in each Brazilian state capital that would work together to sequence 100 samples of the COVID-19 virus every month.
“The funding arrived in November, and we have since hired coordinators, received the first reagents, and started training staff,” says Sabino. The initiative is currently operating in 19 state capitals, including states such as Amapá, Acre, and Rondônia, which are far from the country’s economic centers in the south and are rarely included in large-scale projects due to a lack of infrastructure, explains the scientist. Some laboratories are already carrying out genome analyses. The initial agreement with the business group is set to last for six months.
Adenir Britto / PMSJCCOVID-19 test: low number of RT-PCR molecular tests makes genomic surveillance difficultAdenir Britto / PMSJC
“This initiative by Mulheres do Brasil is important because it is educating groups that have no experience with sequencing,” emphasizes Renato Santana, a virologist from the Department of Genetics, Ecology, and Evolution at the Institute of Biological Sciences of the Federal University of Minas Gerais (ICB-UFMG) whose laboratory is part of the COVID-19 Omics Network. “They are receiving the equipment and supplies needed to sequence and monitor SARS-CoV-2 in their own locations.”
The laboratory headed by Santana will also receive reagents from NVAP, as will the laboratories used by the Mulheres do Brasil project and others from the COVID–19 Omics Network. “The goal is to sequence at least 5,000 samples per month in the five regions of Brazil,” says the scientist.
One of the project’s most important partners, says Santana, is the Hermes Pardini group, based in Belo Horizonte, which has more than 6,000 clinical analysis laboratories nationwide. “All 16,000 positive COVID samples diagnosed by the group since January this year are already in my laboratory. We are just waiting for the reagents to arrive before we redirect them to the laboratories that will carry out the sequencing,” she says.
BRICS Surveillance
The lack of samples to sequence in Brazil is a challenge for those working in genomic surveillance, as explained by Ana Tereza Ribeiro de Vasconcelos, head of the Bioinformatics Laboratory (Labinfo) at the MCTI’s National Laboratory for Scientific Computing (LNCC). “It has been difficult to obtain samples. Testing rates are low now—most people have stopped doing RT-PCR molecular tests, which is how samples of the virus are collected, instead doing antigen tests in pharmacies or at labs, which do not yield any samples,” says the researcher.
Vasconcelos is head of another genome surveillance network that is part of an international cooperation with the other BRICS countries (Russia, India, China, and South Africa). The collaboration, which began in 2021 and is expected to end at the end of this year, was established with the aim of sequencing 3,000 samples in each of the five nations, exchanging information, and performing joint analyses. Another Brazilian initiative is run by the Todos pela Saúde Institute, funded by the Itaú Unibanco Foundation, which invested R$200 million in genome surveillance in 2021.
“I am now sequencing samples collected between January and March that I received from partner laboratories,” says Vasconcelos. The genomes produced by this program in Brazil are shared on GISAID and China’s database, the latter of which is among the three most important in the world. The general coordinator of the project is Brazilian bioinformatics specialist Tulio de Oliveira, who manages the laboratories at the Center for Epidemic Response and Innovation (CERI) at the University of Stellenbosch and the Kwazulu-Natal Research Innovation and Sequencing Platform (KRISP), both in South Africa.
“This virus mutates very quickly and is highly transmissible,” says Vasconcelos. “It is difficult to synchronize the waves of infection that are occurring in different parts of the world. But we are following what happens in each country, and we participate in monthly meetings with scientists from other countries to talk about which variants are circulating in which places. This work is important and can give us an advantage in the fight against the virus.”