In early 2019, scientists from the USA’s Food and Drug Agency (FDA) were surprised by tests of a new bioinformatics methodology that uses computer techniques in biology research. Almost by chance, they discovered that two bulls cloned a few years earlier by bioengineering company Recombinetics, whose cell lines had been genetically edited so that the animals had no horns, were actually transgenic. Before this, nobody had known that the genome of these bulls contained genetic material from another species—more specifically, from the plasmid used to edit their genes. Plasmids are small DNA molecules of bacterial origin used as vectors to insert genetic material into various cell types.
The American authorities considered the finding so important that they immediately published the results on a preprints server, providing access to a version of the paper before peer review. The text was posted in July 2019, and at the time of writing, was awaiting publication in the journal Nature Biotechnology. “Our findings showed where to look for unintended effects of gene editing that scientists may have been missing thus far,” Heather Lombardi, director of the Division of Animal Bioengineering and Cellular Therapies at the FDA’s Center for Veterinary Medicine, told Pesquisa FAPESP (see interview). “Knowing that an entire plasmid can—and sometimes does—enter the animal’s genome will help to improve the quality of the research and the products produced.”
– Gene scissors
– Graham Dellaire: Science with caution
– Heather Lombardi: Support for gene editing
The way the discovery was made almost seems unbelievable. In 2018, the FDA was in the process of developing a method for analyzing next-generation sequencing (NGS) data—a technique that identifies the nitrogen base order of billions of DNA molecules simultaneously—for use in regulatory decisions. The study was conducted by the FDA’s Center for Veterinary Medicine, which needed existing data to validate the methodology. It chose to use data from a study published in Nature Biotechnology in 2016 on the genome sequencing of two cattle bred from cells that had been edited in 2013.
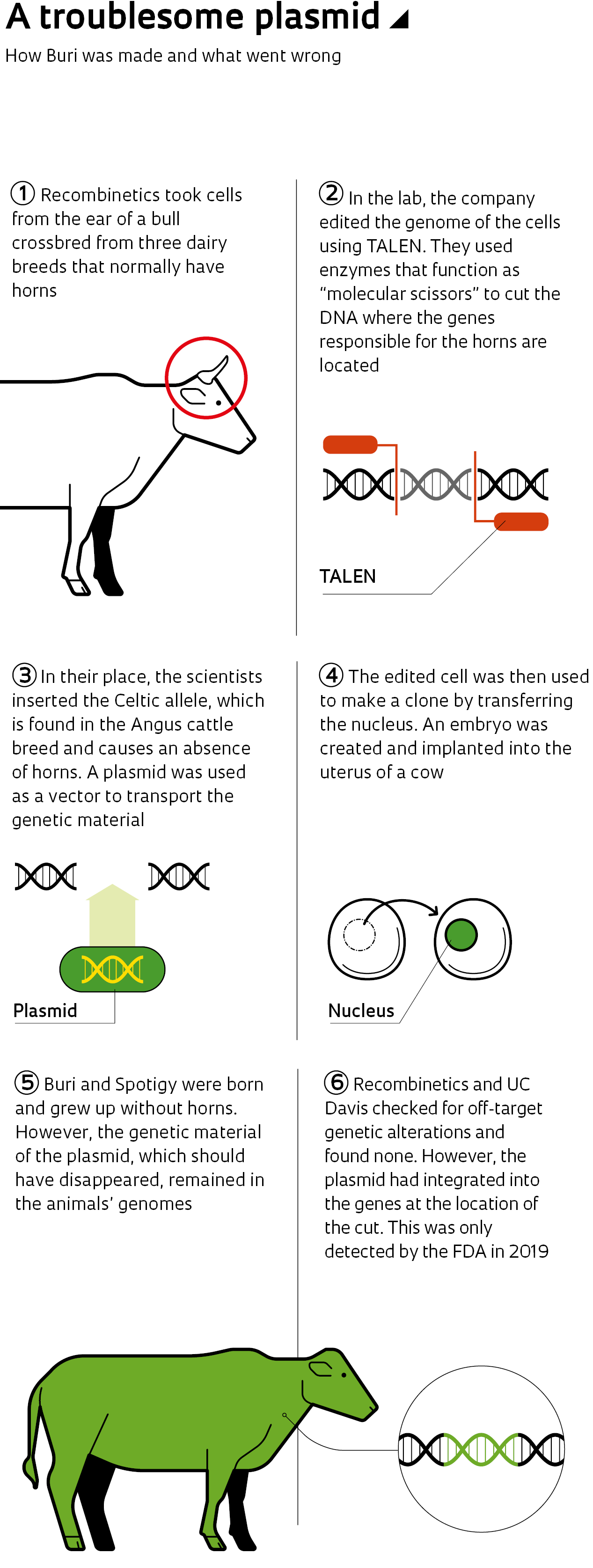
The article, written by American researcher Daniel F. Carlson and colleagues, described the successful gene-editing procedure performed on animals by Recombinetics, a company based in Minnesota (USA). The result was two bulls crossbred from dairy breeds that usually have horns, but genetically edited to grow up hornless. Genetic variants associated with the absence of horns are common in beef cattle, but rare in dairy breeds. The creation of hornless dairy cattle is seen as a major commercial opportunity, since horns make the animals more difficult to manage and producers often choose to dehorn them, a process that is costly, delicate, and painful for the animal.
Genetic editing: a group of technologies that gives scientists the ability to alter the DNA of an organism. It is possible to add, remove, or change genetic material at specific points in the genome
TALEN: a gene editing tool that functions like molecular scissors, cutting the DNA at a specific location
CRISPR-Cas9: a faster, cheaper, and more accurate tool than TALEN. It was adapted from the defense system that naturally protects bacteria from viral infections
Genetically modified organism (GMO): an organism whose genetic material (DNA/RNA) has been modified by a genetic engineering technique
Transgenics: a process through which the genetic material of an organism is changed by adding one or more gene sequences from a different species
New Breeding Technologies (NBTs): set of methodologies and approaches that differ from transgenics in that the modified organism contains no recombinant DNA or RNA
“As our process was designed to detect alterations in samples of genome-edited DNA, we wanted a validation dataset that had both genome-edited and control samples,” said Lombardi. “Carlson’s dataset was chosen as the validation dataset because it had known results. To our knowledge, it was the only publicly available dataset for genome-edited animals at that time, and it was large and of high quality.”
A lot happened between May 2015, when the two calves entered the world, and 2019. And the developments had repercussions even in Brazil. The animals, called Spotigy and Buri, were both cloned from the same bull at the University of Minnesota. They grew up healthy, just like the control calves whose genomes were not edited. In November 2015, aged about six months, they were taken to the University of California (UC), Davis, for research. Spotigy was euthanized for study, while Buri had his semen frozen, later fathering six calves via artificial insemination—five males and one female. The offspring were born in September 2017 at UC Davis, from six different cows of the Hereford breed, which naturally have horns. Four of the six calves inherited the bacterial DNA sequence in their genome.
An article analyzing the genotype and phenotype of these animals, which inherited Buri’s hornlessness, was published in October 2019 by the jourrnal Nature Biotechnology. The study was led by Alison L. Van Eenennaam, an Australian researcher based at UC Davis, and was funded by Recombinetics and the US Department of Agriculture.
In the first half of 2018, before the FDA made its discovery, São Paulo–based company AgroPartners Consulting, which provides consultancy in the field of plant and animal genomics, entered into negotiations with Recombinetics to import Buri’s bull semen to Brazil. “The idea was to inseminate some cows, create a case study, and demonstrate the technology in Brazil,” said veterinarian José Fernando Garcia, a partner at AgroPartners and professor at the School of Veterinary Medicine of São Paulo State University (UNESP), Araçatuba campus.
In 2018, AgroPartners submitted a notice of intent to the Brazilian National Biosafety Technical Commission (CTNBIO) regarding importing Buri’s semen for research and the possibility of selling this type of technology to other companies in the future. In January of the same year, CTNBIO had approved a new resolution (RN16) establishing requirements for New Breeding Techniques—which would cover Buri’s genome editing—and differentiating them from genetic engineering by transgenics.
CTNBIO concluded that according to Article 3 of the Brazilian Biosafety Law, Buri’s semen was not a genetically modified organism (GMO). The commission based the decision on the data available at the time, emphasizing that the inserted alteration—silencing the gene responsible for the occurrence of horns—happens naturally in other bovine breeds.

Alison Van Eenennaam/University of California-Davis
Princess, daughter of Buri (right), at the University of California, DavisAlison Van Eenennaam/University of California-DavisMission aborted
AgroPartners was on the verge of importing the semen when the FDA announced its discovery. “The São Paulo–based company learned of the FDA’s findings and asked CTNBIO to cancel the process, informing them that the bull’s semen was not going to be imported,” says veterinarian Maria Lúcia Zaidan Dagli, a professor at the School of Veterinary Medicine and Animal Science at the University of São Paulo (USP) and a rapporteur at CTNBIO, where she is vice president.
According to Garcia, Buri’s semen was never sent to Brazil. In the USA, five of the six descendants of the genetically modified bull have been euthanized. Only the female, called Princess, whose genome inherited the sequence with the bacterial DNA, remains alive, for research purposes. “She is being bred with a Hereford bull and we hope she is already pregnant,” says Van Eenennaam of UC Davis. “We plan to test the milk to analyze its composition, as suggested by the FDA. Both Princess and her calf will be treated as new animal drugs, an FDA classification that means they will not be able to enter the food chain. They will later be euthanized and cremated.”
The research and sale of transgenic organisms generally follows strict regulatory standards that differ from country to country. According to the World Health Organization (WHO), although the debate encompasses various aspects of transgenics, there are three key concerns: potential allergenic effects of these organisms; potential transfer of genetic material, especially antibiotic resistance genes; and the risks of uncontrolled crossbreeding with nontransgenic organisms.

US Food and Drug Administration
FDA laboratory: agency found that Buri’s genome included genetic material from the plasmidUS Food and Drug AdministrationEditing with talen
To edit the genome of bull cell lines, Recombinetics used a technique known as TALEN, which stands for Transcription Activator-Like Effector Nuclease. Another widely adopted editing technique is CRISPR-Cas9, which is simpler, faster, and cheaper (see report). Both allow scientists to alter a targeted gene or specific region of the genome by deactivating certain genes or inserting modifications.
“Both function like molecular scissors, cutting the DNA,” explains biologist Ângela Saito, a researcher at the Brazilian Center for Energy and Materials Research (CNPEM). “The difference is that TALEN uses modified proteins to identify a specific string of DNA, while CRISPR targets the string using an RNA [ribonucleic acid] molecule, which is faster and simpler to construct than ‘engineering’ a protein.”
Both TALEN and CRISPR-Cas9 can use plasmids as base models to carry genetic instructions. Although plasmids are a common tool in biotechnology laboratories, many researchers prefer to synthesize a model organism that will degrade, rather than remaining in the host and potentially being incorporated into its DNA.
“In 2013, we had to use a plasmid vector because there was not yet any known way to make an exact copy of the Celtic allele [a 212-base-pair insertion and a 10-base-pair deletion],” Daniel Carlson, scientific director at Recombinetics, told Pesquisa FAPESP regarding the bovine genome editing that ensured the absence of horns. “In 2014, when we screened the edited cells, the lab didn’t search directly for the plasmid. Integration of a plasmid model was—and still is—considered a rare event, as supported by the scientific literature.”
Neither Recombinetics nor UC Davis specifically checked whether the plasmid had been integrated before the FDA’s discovery. The unexpected change in Buri’s genome occurred in one of the two alleles of the gene responsible for the presence or absence of horns. In the other allele, the change occurred in the way that scientists predicted and wanted. Every gene that determines a phenotype (a characteristic) has two alleles, which can be identical (homozygous) or different (heterozygous). If they are not identical, the dominant allele determines the displayed characteristic.
With the dominant Celtic allele, Buri and his sibling had no horns. Even Buri’s offspring, the result of crossbreeding with Hereford cows, which normally have horns, grew up to be hornless. The other allele of the same gene, however, unexpectedly integrated the bacterial DNA.
In general, plasmids used as vectors in molecular biology labs contain genes that confer antibiotic resistance, and this was the case with the genetic material integrated by Buri. Experts from Recombinetics, UC Davis, CTNBIO, and AgroPartners do not believe this is a problem. “The plasmid fragments contain bacterial genes that do not function in a mammal. The genes only work in the bacteria, which recognizes the gene as its own and makes it resistant to antibiotics,” says Garcia, from AgroPartners. “In cattle, the gene is an inert fragment. The code is different, it lacks the mechanisms needed in the promoter region for gene expression.”
Veterinarian Marcelo Demarchi Goissis, a researcher at USP’s School of Veterinary Medicine and Zootechnics, agrees with Garcia, but has some reservations: “In theory, this gene does nothing, but this is biology—we know that biology often behaves unexpectedly,” he says. “Initially, there may be no problem, but in an ideal world, the plasmid DNA would not have been incorporated into the bovine genome, because it is difficult to predict what might happen in an entire population of animals.”
A report on the subject in American magazine MIT Technology Review, published by the Massachusetts Institute of Technology (MIT), says it is not clear whether the presence of bacterial DNA necessarily leads to a greater risk. Although the plasmid is unlikely to affect the cattle itself or people who eats its meat, the concern is that the antibiotic resistance gene will be passed onto one of the billions of bacteria present in the body of the bull or cow. Its presence in a cow could create unpredictable opportunities for it to spread, microbiologist John Heritage, from the University of Leeds, UK, told the magazine.
According to Recombinetics, the company never intended to seek FDA approval to sell the animal in the USA or elsewhere. “Buri’s genetics have no commercial value, so seeking regulatory approval made no commercial sense,” says Carlson. For Van Eenennaam from UC Davis, Buri was a prototype, used to test whether genome editing could result in the absence of horns as a dominant characteristic. The next logical step would be to introduce this modification into a horned dairy bull breed using methods that do not involve a plasmid vector. In the view of José Fernando Garcia, the case was merely a “scientific hitch.” “In our opinion, there is no major problem. There are already other animals whose genomes have been edited, both in the USA and in Brazil, with more soon to be born. These animals and their dossiers will be submitted to CTNBIO in a natural, transparent way, as has always been done thus far.”
Scientific articles
CARLSON, D. F. et al. Production of hornless dairy cattle from genome-edited cell lines. Nature Biotechnology. No. 5, pp. 479–81. 2016.
YOUNG, A. E. et al. Genomic and phenotypic analyses of six offspring of a genome-edited hornless bull. Nature Biotechnology. Oct. 2019.
NORRIS, A. L. et al. Template plasmid integration in germline genome-edited cattle. bioRxiv. Online. July 28, 2019.


